- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2017
- Structural biology
- Identification and structure of a novel photoenzyme, fatty acid photodecarboxylase
Identification and structure of a novel photoenzyme, fatty acid photodecarboxylase
Photoenzymes are a very rare type of enzyme that require photons for their catalytic activity. In search for an enzyme responsible for the synthesis of hydrocarbons in microalgae, a flavoprotein that decarboxylates fatty acids using blue light was identified and a crystal structure in complex with a fatty acid substrate was obtained.
Besides the photosynthetic reaction centres, only two families of proteins with light-driven activity have been described: light-dependent protochlorophyllide reductases, which are involved in chlorophyll synthesis in photosynthetic organisms, and DNA photolyases, which repair UV damage in DNA in many organisms ranging from bacteria to animals and plants. While searching for an enzyme converting fatty acids to hydrocarbons, a member of the glucose-methanol-choline (GMC) family of oxidoreductases was identified and shown to be a light-driven enzyme. This novel photoenzyme was christened fatty acid photodecarboxylase (FAP).
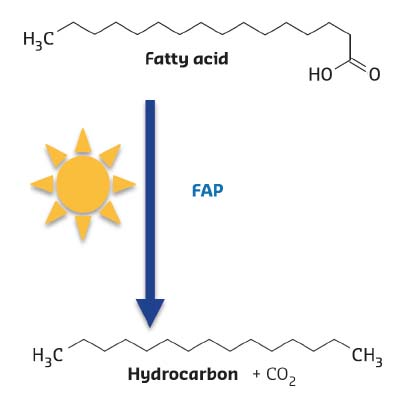
FAP was identified in the green microalgae Chlorella using partial purification of fatty acid decarboxylase activity and proteomic analysis. Expression of the Chlorella gene encoding FAP in the bacterium E. coli resulted in hydrocarbon production, thus confirming its activity. Further biochemical characterisation of the recombinant protein revealed a non-covalently-bound flavine adenine dinucleotide cofactor (FAD), the release of CO2sub> as a co-product and the requirement of constant illumination for activity (Figure 15).
 |
|
Fig. 15: Reaction catalysed by Fatty Acid Photodecarboxylase (FAP). The enzyme is not active in absence of light. |
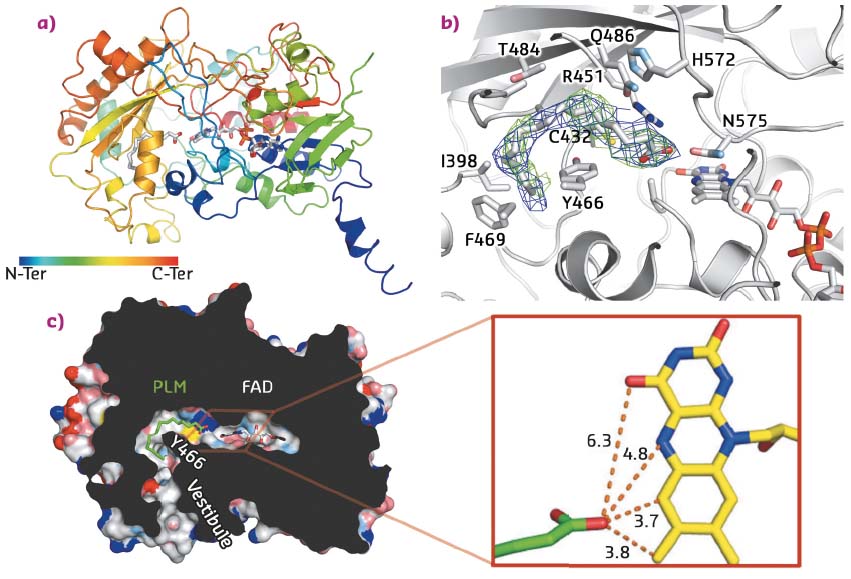
In addition, the protein was successfully crystallised in presence of its fatty acid substrate. X-ray data were collected on the MASSIF-1 ID30A-1 beamline, and the crystal structure solved at 3.15 Å resolution. Despite the fact that the crystals were twinned, the structure was solved by molecular replacement using the 3D structures of domains from two proteins of the GMC oxidoreductase family as models (E. coli choline oxidase and Aspergillus flavus glucose dehydrogenase). Accordingly, the overall architecture of the FAP is broadly similar to that of other GMC oxidoreductases with a typical two-domain fold, the first stabilising the FAD cofactor and the second participating in substrate binding. The crystal structure also revealed a narrow hydrophobic tunnel connecting the FAD cofactor to solvent. Electron density in this tunnel matches with a C16 saturated fatty acid substrate (palmitate) that wraps around the side chain of Y466 (Figure 16). Interestingly, the carboxylate moiety points toward the FAD cofactor. The shortest distance between FAD and the carboxylate is 3.7 Å, suggesting a direct electron transfer between substrate and FAD. Together with a time-resolved spectroscopy study, this structure enabled a mechanism to be proposed in which FAD is excited by blue light to reach an excited state that abstracts an electron from the carboxylate group of the fatty acid substrate stabilised in the hydrophobic tunnel. The radical fatty acid would then decarboxylate spontaneously to yield a hydrocarbon.
 |
|
Fig. 16: Structural features of FAP. a) Overall architecture of the enzyme in complex with FAD and palmitate. b) Details of the palmitic acid binding site with the side chains of residues within 4 Å of the substrate shown as sticks. Omit map electron density associated to palmitic acid is also shown and contoured at 0.5σ (2Fo-Fc; blue) and 2σ (Fo-Fc; green). c) Slicethrough the surface representation of the fatty acid photodecarboxylase in complex with a palmitate substrate (PLM). For clarity, small cavities in the interior of the enzyme are not shown. Inset: Distances (Å) between the carboxyl groups of palmitate and FAD. |
The hydrocarbon-forming microalgal enzyme FAP represents the third enzyme family that uses light directly as an energy source for catalysis. The crystal structure of FAP shows that its overall architecture is related to that of other, thermally-activated GMC oxidoreductase family members despite the unique light-driven decarboxylation activity of the FAP. The crystal structure also suggests residues that may be involved in facilitating the radical-based photochemistry after visible light excitation of the flavin cofactor.
Principal publication and authors
An algal photoenzyme converts fatty acids to hydrocarbons, D. Sorigué (a), B. Légeret (a), S. Cuiné (a), S. Blangy (a), S. Moulin (a), E. Billon (a), P. Richaud (a), S. Brugière (b), Y. Couté (b), D. Nurizzo (c), P. Müller (d), K. Brettel (d), D. Pignol (e), P. Arnoux (e), Y. Li-Beisson (a), G. Peltier (a) and F. Beisson (a), Science 357, 903-907 (2017); doi: 10.1126/science.aan6349.
(a) LB3M, Biosciences and Biotechnologies Institute of Aix-Marseille, CEA, CNRS and Aix-Marseille University, Cadarache (France)
(b) Large Scale Biology Laboratory, CEA and INSERM and University Grenoble Alpes, Grenoble (France)
(c) ESRF
(d) Institute for Integrative Biology of the Cell, CEA, CNRS and Paris-Sud University, Gif-sur-Yvette (France)
(e) LBC, Biosciences and Biotechnologies Institute of Aix-Marseille, CEA, CNRS and Aix-Marseille University, Cadarache (France)



