- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2016
- Structural biology
- Inositol pyrophosphate sensor domains control phosphate homeostasis in eukaryotic cells
Inositol pyrophosphate sensor domains control phosphate homeostasis in eukaryotic cells
SPX domains of previously unknown function are present in fungi, plants and animals. Several SPX domain crystal structures reveal a novel fold and a binding site for inositol pyrophosphates, enigmatic signalling molecules whose levels change in response to nutrient starvation.
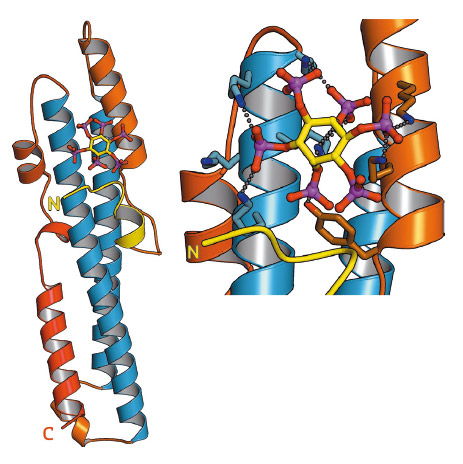
SPX domains are small, soluble domains found in fungi, plants and animals. They can exist as stand-alone modules but are often located at the N-termini of proteins involved in phosphate uptake, transport, storage, metabolism or signalling. We determined 3.3 – 1.9 Å structures of fungal and human SPX domains from crystals obtained by carrier driven crystallisation and including data collected at beamline ID29 [1]. The different structures revealed a new fold with two long a-helices, connected by linkers of variable size. These core helices and two smaller C-terminal helices together form a 3-helix bundle, which is preceded by a flexible N-terminal helical hairpin (Figure 91). Many invariant lysine residues, which represent sequence fingerprints for SPX domains, are clustered in proximity to the N-terminal hairpin structure. A combination of genetic and biochemical experiments in yeast and Arabidopsis revealed that this basic surface represents a docking platform for inositol pyrohosphosphates (PP-InsPs), signalling molecules with poorly characterised cellular functions (Figure 91).
 |
|
Fig. 91: Ribbon representation of the crystal structure of the SPX domain of C. thermophilum glycerophosphodiesterase SPX domain in complex with inositol hexakisphosphate (stick representation). Inset: close-up view of the PP-InsP binding site, with many conserved lysine residues coordinating the phosphate groups of the ligand (dotted lines). |
The concentrations of PP-InsP are known to change in cells, depending on whether they are supplied with sufficient amounts of inorganic phosphate or whether they experience phosphate starvation. We could demonstrate that SPX domains, when bound to PP-InsPs can interact with other proteins. In plants, one such interaction partner is a transcription factor, which is switched on under phosphate starvation to induce expression of genes that allow the plant to respond to the lack of this important nutrient. Under normal growth conditions, when PP-InsP levels are high, the transcription factor is kept inactive by forming a PP-InsP-dependent complex with a plant SPX domain. Under phosphate starvation, the lack of PP-InsP leads to dissociation of the transcription factor – SPX domain complex, thereby enabling the transcription factor to transcribe its target genes.
Taken together, our work defines SPX domains as cellular receptor for PP-InsPs, which control phosphate uptake, transport, storage, metabolism and signalling in fungi, plants and animals.
Principal publication and authors
Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains, R. Wild (a), R. Gerasimaite (b), J.Y. Jung (c), V. Truffault (d), I. Pavlovic (e), A. Schmidt (b), A. Saiardi (f), H.J. Jessen (g,h), Y. Poirier (c), M. Hothorn (a) and A. Mayer (b), Science 352, 986-990 (2016); doi: 10.1126/science.aad9858.
(a) Structural Plant Biology Laboratory, Department of Botany and Plant Biology, University of Geneva (Switzerland)
(b) Department of Biochemistry, University of Lausanne (Switzerland)
(c) Department of Plant Molecular Biology, University of Lausanne (Switzerland)
(d) Department of Biochemistry, Max Planck Institute for Developmental Biology, Tübingen (Germany)
(e) Department of Chemistry and Pharmacy, University of Zürich (Switzerland)
(f) Medical Research Council Laboratory for Molecular Cell Biology, University College London (UK)
(g) Department of Chemistry and Pharmacy, University of Zürich, (Switzerland)
(h) Institute of Organic Chemistry, Albert-Ludwigs-University Freiburg (Germany)
References
[1] R. Wild and M. Hothorn, Protein Sci. 26, 365-374 (2017); doi: 10.1002/pro.3073.



