- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2014
- Structural biology
- Structural basis for nuclear import of splicing factors
Structural basis for nuclear import of splicing factors
The nuclear/cytoplasmic trafficking machinery that is essential for metazoan development and homeostasis, is often deregulated in cancer and can be hijacked by pathogens, such as HIV. The transport of macromolecules between cytoplasm and nucleus is orchestrated by nuclear import and export receptors. Referred to as importins and exportins, these proteins bind their cargos and translocate them across the nuclear pore complex. This process is regulated by Ran, a small GTPase, which partitions between cytoplasm and nucleus in the predominantly GDP- and GTP- bound form, respectively. Importins associate with their cargos in the cytoplasm, and the competitive binding to RanGTP induces them to release their cargos in the nucleus. Most nuclear import/export receptors belong to the β-karyopherin family of proteins of which 22 members encoded in the human genome. Typically, β-karyopherins bind their cargos directly, recognising a linear nuclear localisation or export signal, and/or a specific tertiary/quaternary structural feature. Small molecules targeting nuclear/cytoplasmic trafficking are currently being evaluated as cancer therapeutics.
One fundamentally important type of a nuclear localisation signal, comprising sequences rich in Arg-Ser dipeptides (known as RS domains), belongs to the family of Ser/Arg-rich (SR) proteins. These nuclear proteins also contain RNA recognition motif (RRM) domains and play essential roles in pre-mRNA splicing and 3´-processing, participating in transcription regulation, mRNA transport, translation and nonsense-mediated decay. The canonical splicing factors ASF/SF2 and SC35 are among the best-characterised SR proteins. RS domains are processively phosphorylated on their Ser residues by a set of dedicated kinases. This phosphorylation is thought to trigger import of SR proteins into the nucleus, where it is subsequently required for the spliceosome assembly. During pre-mRNA splicing and 3´-processing, RS domains are involved in establishing networks of protein-protein interactions.
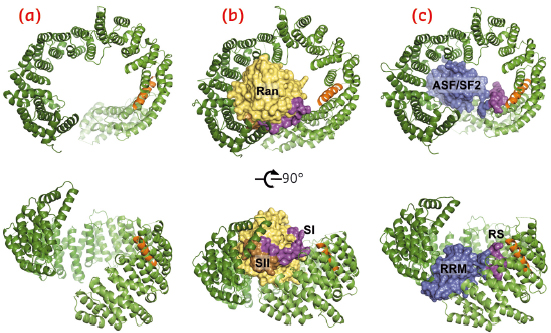
The human β-karyopherin Tnpo3 was shown to recognise the RS domains of ASF/SF2 and SC35 and execute their nuclear import. Likewise, Mtr10, the yeast version of Tnpo3, has long been known to facilitate nuclear import of splicing factors, but no atomic structural information on any of the Tnpo3 orthologs has been reported to date. Using data collected at beamlines ID14-4 (now ID30B) and ID23-1, we determined crystal structures of the human β-karyopherin in its three key function states: as a free protein, and as complexes with RanGTP and ASF/SF2 (Figure 106).
 |
|
Fig. 106: Overview of the crystal structures of unliganded Tnpo3 (a), Tnpo3-RanGTP (b) and Tnpo3-ASF/SF2 (c) complexes. The structures were refined to 3.0, 3.4 and 2.6 Å resolution, respectively. Tnpo3 is shown as cartoons, while ASF/SF2 and Ran are shown in space-filling mode. Tnpo3 is coloured green, except the R-helix, which is shown in orange; Switch I (SI) and Switch II (SII) regions of Ran are shown in magenta and brown, respectively, with the remainder of the Ran structure shown in yellow; RRM2 and RS domains of ASF/SF2 are shown in blue and magenta, respectively. |
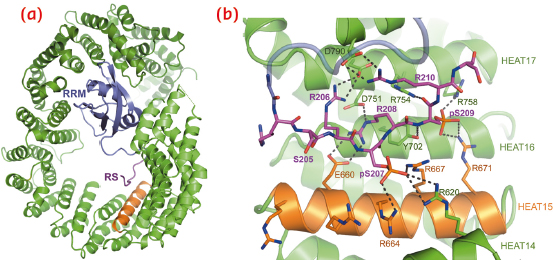
Tnpo3 is a highly flexible molecule, comprised of 20 HEAT repeats. The protein adopts an open circular/toroidal shape, with its N- and C-terminal arches facing each other (Figure 106a). The structure of the Tnpo3-ASF/SF2 complex revealed a tripartite protein-protein interface (Figure 106c, 107a), with the most important contacts involving the RS domain of the cargo. In its mode of binding to Tnpo3, the RS domain is reminiscent of Velcro tape, and its structure is then determined by the underlying Tnpo3 surface. Therefore, the interface is highly adaptable, aiding the β-karyopherin to accommodate highly degenerate RS domains. The inner helix of Tnpo3 HEAT repeat 15 (R-helix) projects an Arg side chain at each turn towards the concave surface of Tnpo3 and contributes to the phospho-Ser binding platform (Figure 107b). The phosphorylated RS domain forms a tight network of salt bridges with complementary Tnpo3 residues on and around the R-helix (Figure 107b).
 |
|
Fig. 107: Details of the structure of Tnpo3-ASF/SF2. a) Overview with both protein chains displayed as cartoons. The RRM2 and RS domains of ASF/SF2 and the R-helix of Tnpo3 are indicated. b) Details of the interface between Tnpo3 and the RS domain seen in the structure of ASF/SF2. |
The flexibility of Tnpo3 may allow it to bind a wide range of SR proteins and, furthermore, aid in their release upon engaging RanGTP in the nucleus. A close comparison of the structures of Tnpo3-ASF/SF2 and of Tnpo3-RanGTP (Figure 106b) revealed that the binding platforms for RanGTP and ASF/SF2 overlap on the concave surface of Tnpo3, with the Switch I region of Ran obstructing the approach of the RS domain to the R-helix. Collectively, these observations provide the structural basis for the dissociation of Tnpo3-cargo complexes via competitive binding of RanGTP upon nuclear entry.
Principal publication and authors
G.N. Maertens (a), N.J. Cook (b), W. Wang (c), S. Hare (a), S.S. Gupta (a), I. Öztop (c), K.-E. Lee (d), V.E. Pye (b), O. Cosnefroy (b), A.P. Snijders (b), V.N. KewalRamani (b), A. Fassati (e), A. Engelman (c) and P. Cherepanov (a,b), PNAS 111, 2728–2733 (2014).
(a) Division of Infectious Diseases, Imperial College London (UK)
(b) Clare Hall Laboratories, Cancer Research UK, London Research Institute, Potters Bar (UK)
(c) Department of Cancer Immunology and AIDS, Dana–Farber Cancer Institute, Boston (USA)
(d) HIV Drug Resistance Program, National Cancer Institute, Frederick (USA)
(e) Wohl Virion Centre and Medical Research Council Centre for Medical and Molecular Virology, Division of Infection and Immunity, University College London (UK)



