- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2005
- Structural Biology
- Crystal Structure of Drosophila Period Protein: Insight into Circadian Clock Regulation
Crystal Structure of Drosophila Period Protein: Insight into Circadian Clock Regulation
Most organisms exhibit 24-hour activity cycles called circadian rhythms (from the latin circa diem = about a day). Circadian rhythms are generated by circadian clocks, which are operated by interconnected transcriptional-translational feedback loops [1]. Period (PER) proteins are central components of the Drosophila and mammalian circadian clocks. Their function is controlled by daily changes in synthesis and degradation, cellular localisation, phosphorylation as well as specific interactions with other clock components. In order to shed some light on these molecular mechanisms, we have solved the crystal structure of a Drosophila Period (dPER) fragment comprising two tandemly-organised PAS (PER-ARNT-SIM) domains (PAS-A and PAS-B) and two additional C-terminal ![]() -helices (
-helices (![]() E and
E and ![]() F). To this end, we have collected 3.5 Å SeMet-MAD data for experimental phasing at beamline ID29 and a 3.15 Å native data set for final refinement at beamline ID14-2.
F). To this end, we have collected 3.5 Å SeMet-MAD data for experimental phasing at beamline ID29 and a 3.15 Å native data set for final refinement at beamline ID14-2.
 |
|
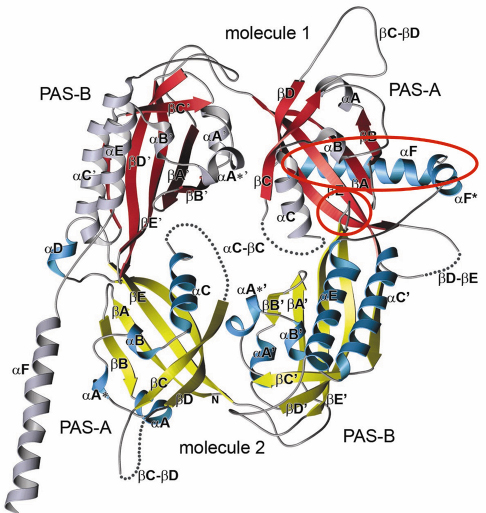
Fig. 79: Crystal structure of the Drosophila Period (dPER) dimer. Molecule1 is shown in red and grey, molecule2 in yellow and blue. PAS-A and PAS-B denote the two PAS domains in each monomer. The PAS-A dimer interfaces are highlighted by red circles. |
PAS domains represent a diverse and ubiquitous family of sensory-, signalling- and protein-protein interaction modules, which consist of a five-stranded antiparallel ß-sheet (ßA-ßE) covered on one face by several a-helices (![]() A-
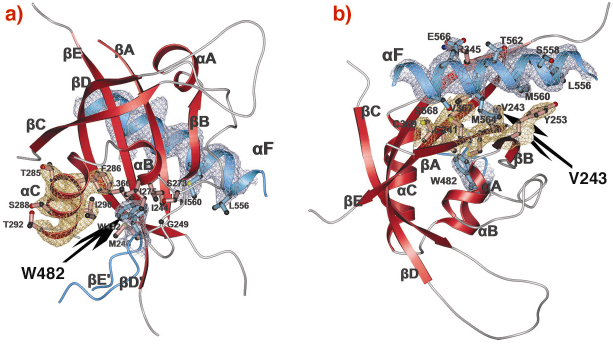
A-![]() C). The crystal structure of the dPER PAS repeat region reveals a non-crystallographic dimer stabilised by two sets of intermolecular PAS domain interactions (Figure 79). The dimer interfaces are mediated by interactions of PAS-A with a conserved tryptophan residue, Trp482, in the ßD’-ßE’-loop of PAS-B (PAS-A-Trp interface, Figure 79 and Figure 80a) and with the helix
C). The crystal structure of the dPER PAS repeat region reveals a non-crystallographic dimer stabilised by two sets of intermolecular PAS domain interactions (Figure 79). The dimer interfaces are mediated by interactions of PAS-A with a conserved tryptophan residue, Trp482, in the ßD’-ßE’-loop of PAS-B (PAS-A-Trp interface, Figure 79 and Figure 80a) and with the helix ![]() F located at the C-terminal to PAS-B (PAS-A-
F located at the C-terminal to PAS-B (PAS-A-![]() F interface, Figures 79, 80b), respectively. Interestingly, a point mutation which leads to extended (29 hour) days in living flies (perL mutation) is located in the centre of the PAS-A-
F interface, Figures 79, 80b), respectively. Interestingly, a point mutation which leads to extended (29 hour) days in living flies (perL mutation) is located in the centre of the PAS-A-![]() F interface. The perL mutation, which corresponds to an exchange of Val243 to Asp (V243D), dissociates the dPER dimer in gel filtration analysis, presumably by introducing a negative charge into this hydrophobic interface (Figure 80b). Thus we propose that the PAS-A-
F interface. The perL mutation, which corresponds to an exchange of Val243 to Asp (V243D), dissociates the dPER dimer in gel filtration analysis, presumably by introducing a negative charge into this hydrophobic interface (Figure 80b). Thus we propose that the PAS-A-![]() F interaction plays a critical role in circadian clocks and that the 29h phenotype of perL mutant flies is caused by a destabilisation of this interface.
F interaction plays a critical role in circadian clocks and that the 29h phenotype of perL mutant flies is caused by a destabilisation of this interface.
 |
|
Fig. 80: PAS-A dimer interfaces of dPER. The PAS-A domain of molecule 1 is shown in red. Helix |
Site-directed mutagenesis studies, guided by our structure, have allowed us to document the functional significance of both dPER dimer interfaces. The strong conservation of Trp482 in mammalian PER (mPER) homologues suggests, that the PAS-A-Trp interface is also present in the mPER dimers observed in the mammalian circadian clock. Additionally, our structure has led us to propose a model for dPER interaction with Timeless, the second central component of the Drosophila clock. Future structural analysis and structure-based biochemical experiments should improve our understanding of the molecular mechanisms underlying circadian clock regulation.
References
[1] N. Cermakian and P. Sassone-Corsi, Nature Reviews, 1, 59-67 (2000).
Principal Publication and Authors
Ö. Yildiz (a), M. Doi (b), I. Yujnovsky (b), L. Cardone (b), A. Berndt (a), S. Hennig (a), S. Schulze (a), C. Urbanke (c), P. Sassone-Corsi (b) and E. Wolf (a), Mol. Cell 17, 69-82 (2005).
(a) Max-Planck-Institute for Molecular Physiology, Department of Structural Biology, Dortmund (Germany)
(b) Institute de Génétique et de Biologie Moléculaire et Cellulaire, Strasbourg (France)
(c) Medizinische Hochschule Hannover, Biophysikalisch-Biochemische Verfahren, Hannover (Germany)



