- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2004
- Structural Biology
- The Structure and Receptor Binding Properties of the 1918 Influenza Haemagglutinin
The Structure and Receptor Binding Properties of the 1918 Influenza Haemagglutinin
An estimated 20 million people died in the influenza pandemic of 1918. The virus responsible probably originated from an avian virus that was introduced into the human population. The ability to be passed from human to human, coupled with a novel antigenicity, was a crucial factor in this virus being able to cause such devastation. Haemagglutinin (HA) is a surface glycoprotein of Influenza A viruses that mediates both viral binding to host cells and the subsequent membrane fusion required for virus invasion. Antibodies that block these activities neutralise virus infectivity. So HA plays an important role in defining the characteristics of the 1918 virus. Therefore, we have structurally characterised the 1918 HA to investigate features of the molecule that may have contributed to the explosive nature of the 1918 pandemic. We have also determined the structures of HA from the first swine influenza virus that was isolated in 1930, and from one of the first human influenza viruses that was isolated in 1934.
There are 15 antigenic subtypes of HA (H1-H15) all of which are found on influenza viruses that infect avian species. Only three human influenza viruses have caused pandemics in the last century and each contained a different HA (H1 in 1918, H2 in 1957 and H3 in 1968).
The cellular receptors that HAs recognise are sialic acids linked to cell-surface glycoproteins and glycolipids. All subtypes of HA found in avian species prefer binding to sialic acid in an ![]() 2,3-linkage to galactose. In contrast, the HAs of human viruses recognise sialic acid in
2,3-linkage to galactose. In contrast, the HAs of human viruses recognise sialic acid in ![]() 2,6-linkage. As a consequence, the cross-species transfer of avian viruses into humans that results in pandemics requires a change in binding specificity.
2,6-linkage. As a consequence, the cross-species transfer of avian viruses into humans that results in pandemics requires a change in binding specificity.
The mechanism that human viruses have used to achieve these changes appears to be different for different subtypes. For the HAs of the H2 and H3 human viruses a minimum of two changes in binding site amino acids, Gln-226->Leu and Gly-228->Ser, correlate with the shift from binding avian to binding human receptors. In contrast, HAs of human H1 viruses (including the 1918 virus) acquire binding to human receptors whilst retaining Gln-226 and Gly-228.
To analyse the mechanisms of receptor binding and the structural basis of receptor specificity in the H1 subtype we determined the structures of ![]() 2,3- and
2,3- and ![]() 2,6-linked sialyl pentasaccharides as analogues of avian and human receptors respectively, in complex with the 1934 human HA and the 1930 swine HA. We then compared these structures to the 1918 HA (Figure 73).
2,6-linked sialyl pentasaccharides as analogues of avian and human receptors respectively, in complex with the 1934 human HA and the 1930 swine HA. We then compared these structures to the 1918 HA (Figure 73).
 |
|
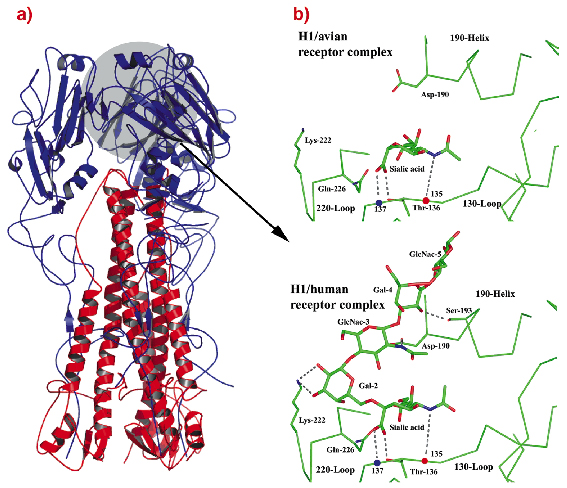
Fig. 73: a) Ribbons diagram of the trimer of 1918-human HA, coloured blue and red for the two polypeptides that form each subunit. b) 1930-swine HA receptor-binding site in complex with avian receptor and with human receptor. Two of the five HAs reported from 1918 viruses have receptor-binding sites indistinguishable from those of 1930-swine HA. The other three 1918 HAs differ only at residue 225. |
Overall, the results suggest that the ability of the 1918 virus to bind human receptors, and thus its ability to spread in the human population, is attributable to the distinct geometry of the 1918 HA receptor binding site and the novel positioning of residues 226 and 228.
By recognizing human receptors, the human-1918 HA fulfils the first requirement of an epidemic virus the ability to spread in the human population. The importance of this requirement is emphasized in the 1997 and 2004 outbreaks of H5 "chicken" influenza in Asia where the virus is extremely virulent for both chickens and humans, but has not acquired the ability to bind ![]() 2,6-linked sialosides and is therefore unable to spread. With the ability to ensure the efficiency of the initial stages of virus infection, coupled with novel antigenicity, the human-1918 HA may alone have been sufficient to generate the high pathogenicity of the 1918 pandemic virus.
2,6-linked sialosides and is therefore unable to spread. With the ability to ensure the efficiency of the initial stages of virus infection, coupled with novel antigenicity, the human-1918 HA may alone have been sufficient to generate the high pathogenicity of the 1918 pandemic virus.
Principal Publication and Authors
S.J. Gamblin (a), L.F. Haire (a), R.J. Russell (a), D.J. Stevens (a), B. Xiao (a), Y. Ha (b), N. Vasisht (a), D.A. Steinhauer (a), R.S. Daniels (a), A. Elliot (a), D.C. Wiley (b) and J.J. Skehel (a), Science 303, 1838-1842 (2004).
(a) Medical Research Council (MRC) National Institute for Medical Research, London (UK)
(b) Howard Hughes Medical Institute, Harvard University, Cambridge, MA (USA)



