- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2008
- Structural biology
- Structural insights into the DegP protease-chaperone
Structural insights into the DegP protease-chaperone
Most proteins acquire a precisely-defined 3D-structure to fulfill their cellular functions. However, this molecular architecture is highly susceptible to various stress conditions such as heat-shock. The resultant misfolding not only impairs protein functionality but also generates molecular aggregates that are extremely toxic for the cell. To survive stress conditions, all organisms employ sophisticated protein-quality-control (PQC) systems in which dedicated chaperones and proteases collaborate closely with each other to reduce the amount of misfolded, aggregation-prone protein species produced.
The heat-shock protein DegP, a member of the HtrA family of proteases, is an essential PQC factor in the cellular envelope of Gram-positive bacteria [1] and exhibits strikingly different properties to those of the functionally-related protease complexes found in the cytosol: (i) Since the periplasm is devoid of nucleotides, DegP has to function without ATP. (ii) DegP can reversibly switch between protease and chaperone functions in a temperature- and substrate-dependent manner [2]. This switch in activity allows an immediate response to environmental changes that are sensed more directly in the periplasm than in the cytosol due to the porous character of the outer membrane. (iii) DegP acts exclusively on misfolded proteins and does not depend on co-factor adaptor or regulatory proteins. The autonomy of DegP is based on an intricate multi-domain organisation comprising a chymotrypsin-like protease domain linked to two C-terminal PDZ domains that contain different binding sites for substrate proteins, allosteric effectors and even cellular membranes.
In the absence of stress conditions, DegP exists as a hexamer (DegP6), a structure that we determined in 2002 using data collected at the ESRF [3]. DegP6 was present in a proteolytically inactive state, in which the loop LA played a central role in stabilising the hexamer as well as in inhibiting the proteolytic activity of an adjacent protomer. Recent data revealed that DegP6 acts as a substrate trap that assembles, upon substrate binding, into the functionally active particles DegP12 and DegP24. Notably, the size of the transformed complexes, 12 or 24mer, depends on the mass and concentration of substrate. Moreover, we could also show that complex formation is transient with DegP recycling into the hexameric state after substrates are chopped up. In contrast to complexes with unfolded model substrates, complexes with co-purified outer membrane proteins (OMPs) were remarkably stable. Interestingly, DegP selectively stabilised folded OMP protomers, whereas unfolded membrane proteins were immediately degraded. The role of DegP in safeguarding OMP precursors through the periplasm was confirmed by the phenotype of a degP deletion strain, where OMP targeting to the outer membrane was significantly impaired.
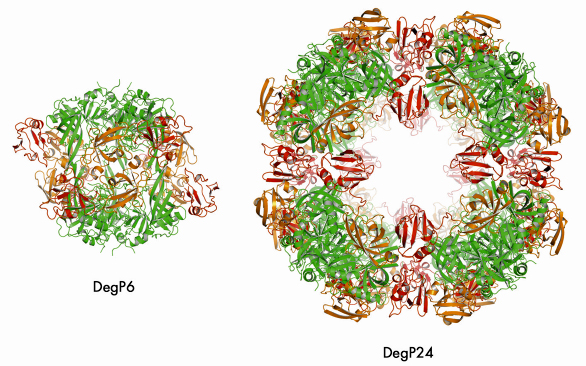
We were able to crystallise the DegP24/OMP(SeMet) complex and performed single anomalous dispersion (SAD) experiments at beamlines ID14-4 and ID23-1. In the resulting structure (Figure 71), eight DegP trimers are located at the vertices of a cube yielding a particle with 432 symmetry. Most importantly, oligomer assembly is correlated with the removal of the inhibitory loop LA from the active site yielding a fully functional DegP protease. Unfortunately, we could not observe the bound OMPs in our structure, most likely due to positional and chemical heterogeneity. However, cryo electron microscopy (carried out by E. Schäfer and H. Saibil, Birkbeck College, London) of the DegP12/OMP complex revealed that OMPs are bound - presumably in a folded state - in the inner cavity of DegP thus confirming our biochemical data.
 |
|
Fig. 71: Comparison of DegP6 and DegP24. The colour code is the same for the two molecules: protease domains (green), PDZ1 (orange), PDZ2 (red). |
Taken together, our work has revealed a novel regulatory mechanism for proteases that is based on substrate-induced oligomer assembly and has provided molecular insight into the antagonistic protease and chaperone activities of DegP. While encapsulation of folded OMP protomers is protective and might allow safe transit through the periplasm, misfolded proteins are eliminated in the proteolytic folding chamber.
Principal publication and authors
T. Krojer (a), J. Sawa (a), E. Schäfer (b), H.R. Saibil (b), M. Ehrmann (c) and T. Clausen (a), Nature 453, 885 (2008).
(a) Research Institute for Molecular Pathology - IMP, Vienna (Austria)
(b) Crystallography Department and Institute of Structural Molecular Biology, Birkbeck College, London (UK)
(c) Centre for Medical Biotechnology, FB Biology and Geography, University Duisburg-Essen, Essen (Germany)
References
[1] B. Lipinska, O. Fayet, L. Baird and C. Georgopoulos, Journal of Bacteriology 171, 1574 (1989).
[2] C. Spiess, A. Beil and M. Ehrmann, Cell 97, 339 (1999).
[3] T. Krojer, M. Garrido-Franco, R. Huber, M. Ehrmann and T. Clausen, Nature 416, 455 (2002).



