- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2005
- Structural Biology
- The Myosin VI Structure Reveals the Mechanism of its Reverse Directionality
The Myosin VI Structure Reveals the Mechanism of its Reverse Directionality
Nature has evolved a number of molecular motors which accomplish motility by producing directed force along cytoskeleton filaments. The myosin superfamily regroups the motors that progress along an actin filament by conversion of chemical energy (provided by ATP hydrolysis) into mechanical energy, while transporting cargos and vesicles throughout the cell. The current view of how myosins couple ATP hydrolysis and actin binding to movement is known as the “swinging lever arm hypothesis”. Briefly, the motor/ATP hydrolysis cycle is coupled to small movements within the motor domain, that are amplified and transmitted via the “converter” subdomain to an elongated region, called the lever arm. The lever arm (which consists of a target helix and associated light chains) further amplifies the motions of the converter into large directed movements.
Myosin VI, like myosin V, has been described as a dimeric processive motor, taking multiple steps along the actin filament before detaching. However, myosin VI, in constrast to all other characterised myosins, moves toward the minus (-) end of the actin filament [1]. Perhaps because of its reverse directionality, myosin VI has another unusual feature: it takes similarly sized steps to those of myosin V, despite having a much shorter lever arm. This property cannot be explained in the context of the “swinging lever arm” theory since its step size is not proportional to its lever arm length. In addition, myosin VI contains a specific, unique, insert of 39 residues located between the motor domain and the lever arm. Together, this evidence suggests that myosin VI produces force using a mechanism different from that of other myosins.
Electron microscopy maps of the myosin VI motor bound to actin gave the first indication that the unique 39 residue insert was involved in the myosin VI reverse directionality. These maps reveal that an ADP-mediated conformational change in the distal part of the motor occurred in the opposite direction to that observed for other myosins and that the lever arm was directed toward the (-) end of the actin filament in both states [1]. However, molecular engineering studies using myosin V/VI chimera, in which the insert was added or removed, concluded that the motor domain, and not the unique insert, determined the direction of myosin motility. Quite unexpectedly, we have later shown that this unique insert corresponds to a novel target sequence for calmodulin with calcium bound.
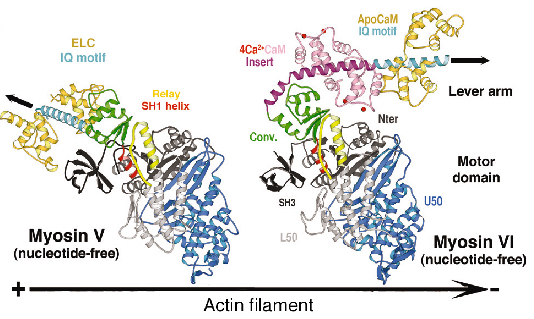
We have determined the structure of myosin VI in a nucleotide-free state at 2.4 Å resolution data collected on ID14, ID23 and ID29 (Figure 81). This structure reveals only minor differences in the structure of the motor domain when compared to myosin V [2]. However the lever arm of myosin VI is directed in the opposite direction towards the (-) end of the actin filament and the unique amino acid insert is a helix that plays a crucial role in this reversal of direction. The proximal end of this helix wraps around the converter rather than emerging as a straight helix while the distal part of the insert forms a previously unseen calmodulin-binding motif.
 |
|
Fig. 81: The structure of nucleotide-free myosin VI compared to that of myosin V (left). Note the difference in the position of the lever arm (IQ motif, cyan) due to the unique insert (purple) and its associated-calmodulin molecule. The convertor domain is highlighted in olive. |
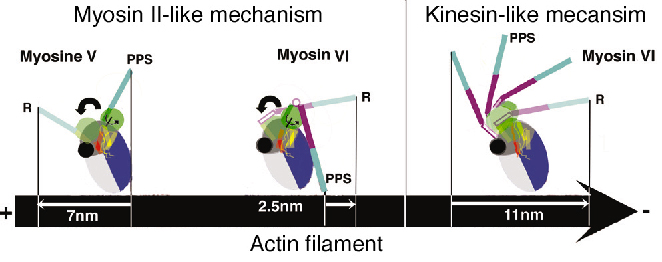
Both the insert and its associated calmodulin make specific interactions with the converter domain. The net result of these interactions is that the lever arm is rotated about 120° from the position of the lever arm found for (+) end motors. This structure, which represents that at the end of the powerstroke, largely accounts for myosin VI reverse directionality. In redirecting the lever arm, the insert creates a bias for the binding of the second head towards the (-) end of actin filament. Modeling of the myosin VI conformation at the beginning of the stroke using the motor domain of myosin II in its pre-powerstroke state reveals that the stroke produced would be much smaller than that experimentally measured. In contrast, similar modeling for myosin V fully accounts for its 7 nm stroke (Figure 82).
 |
|
Fig. 82: Reversal of directionality mechanisms in myosins. Light chains are omitted for clarity. (R) Rigor state: a nucleotide-free high affinity state for actin filament following force production. (PPS) Pre-powerstroke state: an ADP.Pi state of weak affinity for actin filament that preceeds force production. |
Myosin VI thus produces force using a different mechanism to myosin V and we propose that a kinesin-like uncoupling/docking mechanism could fully explain the movements of myosin VI. In this hypothesis, a specific region (the SH1 helix) unfolds to uncouple the lever arm from the motor domain prior to force generation. Its refolding upon force production would impose a stroke that could be as large as 11 nm (Figure 82). However, to further understand how myosin VI works, the structure of this motor in other conformations is needed.
References
[1] A.L. Wells, et al. Nature. 401, 505-508 (1999).
[2] P-D. Coureux, et al. Nature 425, 419-423 (2003).
Principal Publication and Authors
J. Ménétrey (a), A. Bahloul (a), A.L. Wells (b), C.M. Yengo (b), C.A. Morris (b), H.L. Sweeney (b), A. Houdusse (a). Nature, 435, 779-85 (2005).
(a) Structural Motility, Institut Curie CNRS, UMR144, Paris (France)
(b) Department of Physiology, University of Pennsylvania School of Medicine (USA)



