- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2002
- Macromolecular Crystallography
- Structure of the Bovine Lysosomal alpha-Mannosidase, the Enzyme Involved in the Lysosomal Storage Disease alpha-Mannosidosis
Structure of the Bovine Lysosomal alpha-Mannosidase, the Enzyme Involved in the Lysosomal Storage Disease alpha-Mannosidosis
Lysosomal ![]() -mannosidase (LAM) is a member of the glycosyl hydrolase family GH38. It hydrolyses all known
-mannosidase (LAM) is a member of the glycosyl hydrolase family GH38. It hydrolyses all known ![]() -mannosidic linkages in lysosomes. Lysosomes are cellular particles, which are responsible for the breakdown of cellular end products. They have an internal pH of 4.5 and are thus much more acidic than the rest of the cell. Errors in lysosomal processes lead to a number of inherited lysosomal storage diseases, many of which are very serious. These include
-mannosidic linkages in lysosomes. Lysosomes are cellular particles, which are responsible for the breakdown of cellular end products. They have an internal pH of 4.5 and are thus much more acidic than the rest of the cell. Errors in lysosomal processes lead to a number of inherited lysosomal storage diseases, many of which are very serious. These include ![]() -mannosidosis, which is a rare disease in humans, cattle, cats and guinea pig. Lack of LAM activity causes swelling of the lysosomal vacuoles, and apparently leaking of the unhydrolysed sugars from these vacuoles causes the symptoms of the disease; mental retardation, skeletal changes, hearing loss and reduced immunity in humans [1, 2]. Two protein level
-mannosidosis, which is a rare disease in humans, cattle, cats and guinea pig. Lack of LAM activity causes swelling of the lysosomal vacuoles, and apparently leaking of the unhydrolysed sugars from these vacuoles causes the symptoms of the disease; mental retardation, skeletal changes, hearing loss and reduced immunity in humans [1, 2]. Two protein level  -mannosidosis mutations have been identified in cattle and six in humans.
-mannosidosis mutations have been identified in cattle and six in humans.
We isolated the natural lysosomal ![]() -mannosidase directly from bovine kidneys [2] (10 kg of kidneys yielding 5-20 mg of the enzyme). The LAM protein is a ~250 kDa homodimer which is both glycosylated and proteolytically cleaved during its maturation and transport to lysosomes. The protein was crystallised in a fully-glycosylated form in a large hexagonal unit cell with cell dimensions a = b = 117.88 Å, c = 582.04 Å. Data were collected to 2.7 Å resolution at beamline ID14-4. We solved the structure in P6122 space group by molecular replacement with a distant relative, the Drosophila melanogaster Golgi II
-mannosidase directly from bovine kidneys [2] (10 kg of kidneys yielding 5-20 mg of the enzyme). The LAM protein is a ~250 kDa homodimer which is both glycosylated and proteolytically cleaved during its maturation and transport to lysosomes. The protein was crystallised in a fully-glycosylated form in a large hexagonal unit cell with cell dimensions a = b = 117.88 Å, c = 582.04 Å. Data were collected to 2.7 Å resolution at beamline ID14-4. We solved the structure in P6122 space group by molecular replacement with a distant relative, the Drosophila melanogaster Golgi II ![]() -mannosidase. The asymmetric unit of the crystal contains a single monomer and the crystallographic packing offered two possible LAM dimers. The correct dimer was identified by electron microscopy.
-mannosidase. The asymmetric unit of the crystal contains a single monomer and the crystallographic packing offered two possible LAM dimers. The correct dimer was identified by electron microscopy.
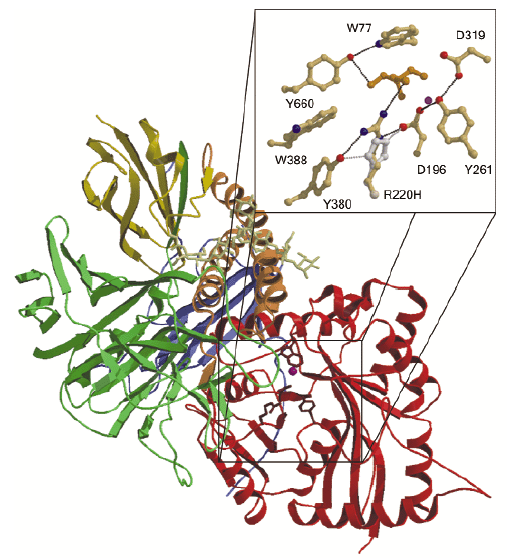
The N-terminal active site domain is a distorted 7-stranded ![]() /ß barrel and the active site is formed on the top of the barrel. Following the barrel domain, the structure consists of a 3-helix bundle, two further small ß domains and a large 17 stranded ß-domain (Figure 12). All known LAMs have a conserved glycosylation site following the 3-helix bundle. In our bovine LAM (bLAM) structure this site contains a high mannose type glycan, which rests against the 3-helix bundle. Two GlcNAc and eight mannose residues are visible in the electron density map. The bLAM structure also contains several salt bridge networks. As these networks are not conserved in related enzymes functioning at neutral pH, it seems likely that these networks are involved in stability and activation of the enzyme at the lysosomal pH (~4.5), which is close to the pKa of aspartic and glutamic acids.
/ß barrel and the active site is formed on the top of the barrel. Following the barrel domain, the structure consists of a 3-helix bundle, two further small ß domains and a large 17 stranded ß-domain (Figure 12). All known LAMs have a conserved glycosylation site following the 3-helix bundle. In our bovine LAM (bLAM) structure this site contains a high mannose type glycan, which rests against the 3-helix bundle. Two GlcNAc and eight mannose residues are visible in the electron density map. The bLAM structure also contains several salt bridge networks. As these networks are not conserved in related enzymes functioning at neutral pH, it seems likely that these networks are involved in stability and activation of the enzyme at the lysosomal pH (~4.5), which is close to the pKa of aspartic and glutamic acids.
 |
|
Fig. 12: The three-dimensional structure of bovine |
Mutations in the LAM amino acid sequence that cause ![]() -mannosidosis in humans are scattered throughout the structure. Only the mutation H72L is in the active site itself, where it affects a metal-coordinating residue. Two others (T355P and P356R) are located at the start of an
-mannosidosis in humans are scattered throughout the structure. Only the mutation H72L is in the active site itself, where it affects a metal-coordinating residue. Two others (T355P and P356R) are located at the start of an  -helix in the active-site domain and presumably disturb the initiation of the helix and folding of the domain. Three further mutations (W714R, R750W and L809P) are located in the 17-stranded ß-domain. Of these, R750W is possibly the most interesting, since it mediates an interaction between three domains.
-helix in the active-site domain and presumably disturb the initiation of the helix and folding of the domain. Three further mutations (W714R, R750W and L809P) are located in the 17-stranded ß-domain. Of these, R750W is possibly the most interesting, since it mediates an interaction between three domains.
Both mutations causing ![]() -mannosidosis in cattle [2] are related to the active site of the enzyme. The R220H mutation in Galloway cattle affects R220, which is hydrogen-bonded both to an important residue in the catalysis, D196, as well as to the substrate mimic Tris. H220 would be able to form hydrogen bonds to D196 and to Y380 as R220 does, but the hydrogen-bonding to Tris and presumably to the substrate would be broken. This mutation will thus most likely affect the substrate binding and also the chemistry of D196. Another mutation, F320L, causes
-mannosidosis in cattle [2] are related to the active site of the enzyme. The R220H mutation in Galloway cattle affects R220, which is hydrogen-bonded both to an important residue in the catalysis, D196, as well as to the substrate mimic Tris. H220 would be able to form hydrogen bonds to D196 and to Y380 as R220 does, but the hydrogen-bonding to Tris and presumably to the substrate would be broken. This mutation will thus most likely affect the substrate binding and also the chemistry of D196. Another mutation, F320L, causes ![]() -mannosidosis in Angus cattle and is also known to affect the stability of the enzyme. In the structure, the aromatic ring in A:F320 stacks against Y84 in a loop which is involved in dimer formation. Its mutation to Leucine would presumably weaken monomer-monomer interactions in the physiological dimer. However, this mutation might also affect catalysis since it follows another active-site residue, D319, in the amino acid sequence.
-mannosidosis in Angus cattle and is also known to affect the stability of the enzyme. In the structure, the aromatic ring in A:F320 stacks against Y84 in a loop which is involved in dimer formation. Its mutation to Leucine would presumably weaken monomer-monomer interactions in the physiological dimer. However, this mutation might also affect catalysis since it follows another active-site residue, D319, in the amino acid sequence.
Our structure determination helps to explain how particular mutations on the LAM amino acid sequence can result in ![]() -mannosidosis and represents the first step in understanding the biological mechanism behind the disease. It also provides a first structure of a mammalian enzyme in this class of glycoside hydrolases and provides an interesting new way of low pH activation.
-mannosidosis and represents the first step in understanding the biological mechanism behind the disease. It also provides a first structure of a mammalian enzyme in this class of glycoside hydrolases and provides an interesting new way of low pH activation.
References
[1] T. Berg, H.M. Riise, G.M. Hansen, D. Malm, L. Tranebjaerg, O.K. Tollersrud and Ø. Nilssen, Am. J. Hum. Genet. 64, 77-88 (1999).
[2] O.K. Tollersrud, T. Berg, P. Healy, G. Evjen, U. Ramachandran and Ø. Nilssen, Eur. J. Biochem. 246, 410-419 (1997).
Principal Publication and Authors
P. Heikinheimo (a), R. Helland (a), H-K. Schrøder Leiros (a), I. Leiros (a), S. Karlsen (a), G. Evjen (a), R. Ravelli (b), G. Schoehn (b,c), R. Ruigrok (b), O-K. Tollersrud (a), S. McSweeney (b,d) and E. Hough (a).
(a) Universitetet i Tromsø, Tromsø (Norway)
(b) EMBL, Grenoble (France)
(c) IBS, Grenoble (France)
(d) ESRF



