- Home
- Users & Science
- Find a beamline
- Structural biology
- Inhouse Research and Development
- Biology
- Deinococcus radiodurans
Deinococcus radiodurans
Structural Biology of Deinoccocus radiodurans
Background
D. radiodurans is a gram-positive bacteria, which exhibits significant resistance to ionising radiation. It is capable of withstanding acute irradiation doses of 1 500 kilorads, a level hundreds of times higher than most other organisms. In most organisms, exposure to such doses of ionising radiation causes massive damage to genomic DNA, however D. radiodurans is capable of surviving and repairing hundreds of double strand breaks without loss of vitality. In comparison, in E. coli five double strand breaks are invariably fatal. The publication of the annotated sequence of the D. radiodurans genome in 1999 (White et al., Science, 286, 1571-1577, 1999) has allowed a much more detailed analysis of the composition of this organism. Surprisingly, despite this analysis, the mechanisms of resistance remain unclear, although some tantalising glimpses of the possible reasons for the resistance have been revealed.
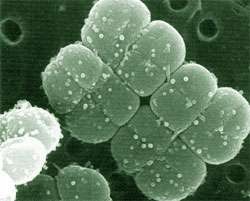
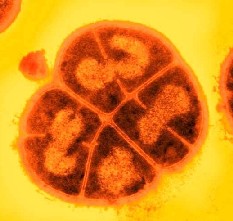 Specific expansion of some protein families has been observed during the analysis of several bacterial genomes. In many cases there appears to be some connection between the protein families and the habitat of the bacteria. D. radiodurans possesses a number of families that are expanded significantly with respect to other bacteria. The families include both characterised and uncharacterised proteins, and most of those to which function can be assigned seem to be related to stress response and the elimination of radiation damage products. The two most prominent family expansions are the Nudix (MutT) family of pyrophosphohydrolases and the DinB family of metal-dependent enzymes of unknown function, but induced by DNA damage. Interestingly, many of these proteins exhibit unusual or unique domain compositions that may indicate novel cellular roles or activity.
Specific expansion of some protein families has been observed during the analysis of several bacterial genomes. In many cases there appears to be some connection between the protein families and the habitat of the bacteria. D. radiodurans possesses a number of families that are expanded significantly with respect to other bacteria. The families include both characterised and uncharacterised proteins, and most of those to which function can be assigned seem to be related to stress response and the elimination of radiation damage products. The two most prominent family expansions are the Nudix (MutT) family of pyrophosphohydrolases and the DinB family of metal-dependent enzymes of unknown function, but induced by DNA damage. Interestingly, many of these proteins exhibit unusual or unique domain compositions that may indicate novel cellular roles or activity.
Analysis of the organisation of the genome of D. radiodurans allows the prediction of those genes that encode proteins that are naturally highly expressed. It is proposed that such behaviour may indicate a protein that is vital to the stability and life cycle of the organism. Such analysis of the D. radiodurans genome reveals four classes of genes, which show significantly enhanced, predicted natural expression levels:
- - degradation and export vehicles for removing damaged DNA, RNA and proteins
- - chaperones, which may enhance the folding and repair of damaged proteins
- - detoxification proteins for the removal of free oxygen radicals and other toxic substances
- - proteases, probably the highest number any of the 61 complete prokaryotic genomes from 48 species
Most known DNA repair proteins are not predicted to be highly expressed, with the exception of the RecA protein, leaving the possibility open that some of the many proteins of, as yet, unknown function, may be involved in novel repair mechanisms.
Together, genomic and bioinformatic data provide a wealth of information that would be greatly enhanced by structural characterisation of some of these proteins. A large question remains as to how it will be possible to determine the large number of structures required in a fast and cost effective manner. Recent results at the ESRF and other synchrotrons suggest that the advantages of third generation SR sources make it feasible to solve a great many structures using only the anomalous scattering from the sulphur atoms that occur naturally in some amino acids. For this technique to be useful all that is required is for the protein under investigation to contain a sufficiently large percentage of cysteine and methionine residues to make the anomalous signal detectable. Analysis of the D. radiodurans genome reveals a significant number of proteins that fit this criterion.
Pilot study, initial results and future extension of the project
In order to investigate the feasibility of a systematic and extensive study of this organism, 100 targets have been cloned in collaboration with the biotech company Protein’eXpert.
The scope of this project can be extended to follow the leads provided by collaborating bioinformatics experts and our own proteomics studies. This may ultimately result not only in a more complete understanding of the radiation resistance of this bacterium, but also to the discovery of novel DNA repair systems, applicable to an understanding of the mechanisms of higher organisms such as man.
People working on these projects

Joanna Timmins

Ana-Maria Goncalves

Olivia Sleator

Structural Biology
Collaborations
PDB codes.
| Id | spaceGroup | A | B | C | Beta | resolution | rfree |
|---|---|---|---|---|---|---|---|
| 1USP | P 1 21 1 | 45.41 | 56.6 | 49.46 | 90.27 | 1.9 | 0.2 |
| 1W3O | C 1 2 1 | 99.86 | 38.95 | 59.81 | 114.25 | 1.6 | 0.21 |
| 1W3P | C 1 2 1 | 99.94 | 38.81 | 60.06 | 114.11 | 1.8 | 0.23 |
| 1w3Q | C 1 2 1 | 99.8 | 38.77 | 60 | 114.05 | 1.88 | 0.22 |
| 1W3R | C 1 2 1 | 99.59 | 39.17 | 59.84 | 114.09 | 1.9 | 0.26 |
| 1W3S | C 1 2 1 | 134.43 | 52.35 | 101.09 | 106.32 | 2.4 | 0.27 |
| 2BHU | P 21 21 21 | 59.58 | 66.62 | 152.51 | 90 | 1.1 | 0.16 |
| 2BHY | P 21 21 21 | 60.03 | 66.56 | 153.14 | 90 | 1.5 | 0.15 |
| 2BHZ | P 21 21 21 | 59.15 | 66.57 | 153.5 | 90 | 1.2 | 0.14 |
| 2BOO | P 21 21 2 | 63.5 | 85.1 | 85.9 | 90 | 1.8 | 0.27 |
| 2BXY | P 21 21 21 | 59.42 | 66.53 | 152.52 | 90 | 1.75 | 0.19 |
| 2BXZ | P 21 21 21 | 59.42 | 66.53 | 152.52 | 90 | 1.75 | 0.19 |
| 2BYQ | P 21 21 21 | 59.51 | 66.65 | 153.1 | 90 | 1.55 | 0.17 |
| 2BY1 | P 21 21 21 | 59.61 | 66.74 | 153.54 | 90 | 1.55 | 0.19 |
| 2BY2 | P 21 21 21 | 59.42 | 66.53 | 152.52 | 90 | 1.5 | 0.18 |
| 2BY3 | P 21 21 21 | 59.51 | 66.65 | 153 | 90 | 1.5 | 0.2 |
| 2C2F | P 2 3 | 90.62 | 90.62 | 90.62 | 90 | 1.61 | 0.15 |
| 2C2J | P 2 3 | 88.45 | 88.45 | 88.45 | 90 | 2.05 | n/a |
| 2C2P | P 6 | 101.73 | 101.73 | 37.56 | 90 | 1.75 | 0.22 |
| 2C2Q | P 6 | 102.49 | 102.49 | 37.58 | 90 | 1.7 | 0.24 |
| 2C2U | P 2 3 | 90.37 | 90.37 | 90.37 | 90 | 1.1 | 0.15 |
| 2C6R | P 2 3 | 88.97 | 88.97 | 88.97 | 90 | 2.1 | n/a |
| 2CDY | P 1 21 1 | 43.58 | 87.1 | 116.42 | 92.1 | 2 | 0.22 |
| 2CE4 | P 1 21 1 | 44.28 | 83.21 | 59.52 | 110.18 | 2.2 | 0.24 |



