- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2016
- Structural biology
- Crystal structure of PolD provides a novel paradigm for the classification of DNA polymerases
Crystal structure of PolD provides a novel paradigm for the classification of DNA polymerases
Polymerase D (PolD), an archaeal replicative DNA polymerase, is the only class of DNA-polymerases whose structure remains unknown and for which the catalytic domain has no assigned fold. The crystal structure of both catalytic subunits of PolD was determined using data collected at beamline ID23-1 and PolD was found to share an unexpected structural homology with the multisubunit transcriptases. This discovery of a common catalytic core suggests that the DNA replication and transcription apparatus may share a joint evolutionary history.
In all forms of cellular life, DNA polymerases (DNAPs) play central roles in genome replication, maintenance, and repair, and have been the subject of intensive research for decades. Over the years, all DNAPs have been grouped into various families, using sequence alignments: PolA, PolB, PolC, PolD, PolX, PolY and reverse transcriptases (RT). Strikingly, nearly all of them belong to one of two different folds, the Klenow-fold (PolA, PolB, PolY and RT), or the Polb-fold (PolC, PolX). The only class of DNA-polymerases left whose structure is unknown and for which the catalytic domain has no assigned fold is PolD.
PolD exists in all Archaea, except Crenarchaea, and is a replicative polymerase responsible for initiating DNA synthesis at both leading and lagging strands. It is composed of a large catalytic subunit (DP2) and a smaller subunit with 3’-5’ proofreading exonuclease activity (DP1). Apart from the N-terminal regions of its DP1 (1-50) and DP2 (50-280) subunits, neither the structure of the catalytic polymerase nor that of the exonuclease domain have yet been determined. While DP1 is known to belong to the calcineurin-like phosphodiesterase superfamily, DP2 shows no sequence similarity to other proteins with the exception of a short C-terminal zinc-binding motif in eukaryotic Pole.
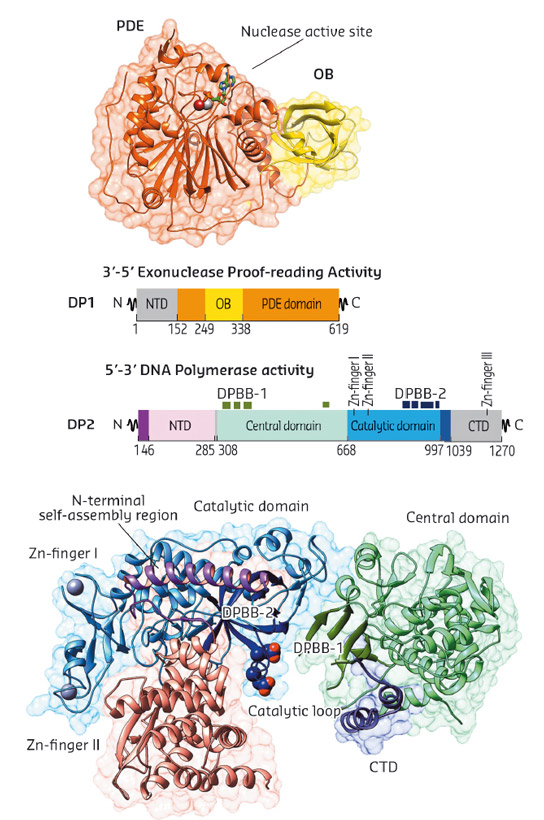
To help resolve the uncertainty concerning the evolutionary origins of D-family DNAP, we determined the crystal structures of two large fragments of both DP1 and DP2 subunits of the Pyrococcus abyssi PolD. Both DP1 and DP2 crystal structures were determined individually using data collected at beamline ID23-1 and refined at final resolutions of 2.5 Å and 2.2 Å, respectively (Figure 84).
 |
|
Fig. 84: Overview of PolD subunit structures. Cartoon representations of DP1 (top) and DP2 (bottom), structures coloured according to domain boundaries. |
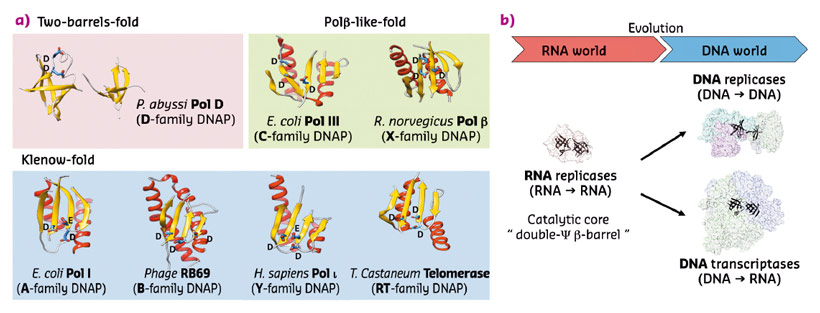
The crystal structures of both DP1 and DP2 subunits reveal that PolD is an atypical DNAP. Firstly, DP1 subunit contains a calcineurin-like phosphodiesterase fold that is responsible for the 3'-5' proof-reading exonuclease activity. However, in most other structurally-characterised DNAPs, the catalytically active proofreading domains fold into an α/β structure with a twisted five-stranded mixed β-sheet, which shares no structural homology with DP1. Secondly, the structure of DP2 catalytic subunit shows that it shares no significant structural similarity with any known DNAP. With knowledge of the structure of PolD, a third structural class of DNA polymerases has been discovered (Figure 85a). In fact, PolD DP2 shares an unexpected structural homology with the "two-barrel" architecture of RNA polymerases (RNAP), which includes multisubunit transcriptases from all domains of life, homodimeric RNA silencing pathway RNAPs and atypical RNAPs encoded by some viruses and phages. This defines a novel paradigm for the classification and possible evolutionary relationships between different types of both RNA and DNA polymerases, which shows, for the first time in the non-viral world, DNA transcription and DNA replication within the same protein superfamily. The finding that PolD and "two-barrel" RNAPs share a common catalytic core with similar sequence motifs suggest that the DNA replication and transcription apparatus may share a joint evolutionary history.
We hypothesise that this "two-barrel" catalytic core common ancestor was present and functional as an RNA replicase in an RNA world. The ability of this catalytic core to acquire novel nucleotide polymerisation activities, like DNA replication and DNA transcription, may have made the RNA-world/DNA-world transition easier. Indeed, the versatility of the "two-barrel" fold may have allowed DNA replication and transcription to jointly evolve from a common catalytic core, rather than being invented separately (Figure 85b).
Principal publication and authors
Shared active site architecture between archaeal PolD and multisubunit RNA polymerases revealed by X-ray crystallography, L. Sauguet (a), P. Raia (a,b), G. Henneke (c,d,e) and M. Delarue (a), Nature Communications 7, 12227 (2016), doi: 10.1038/ncomms12227.
(a) Unit of Structural Dynamics of Macromolecules, Pasteur Institute & CNRS UMR 3528, Paris (France)
(b) Pierre et Marie Curie University, Paris (France)
(c) Ifremer, UMR 6197, Laboratoire de Microbiologie des Environnements Extrêmes, Plouzané (France)
(d) UBO, UMR 6197, Laboratoire de Microbiologie des Environnements Extrêmes, Plouzané (France)
(e) CNRS, UMR 6197, Laboratoire de Microbiologie des Environnements Extrêmes, Plouzané (France)