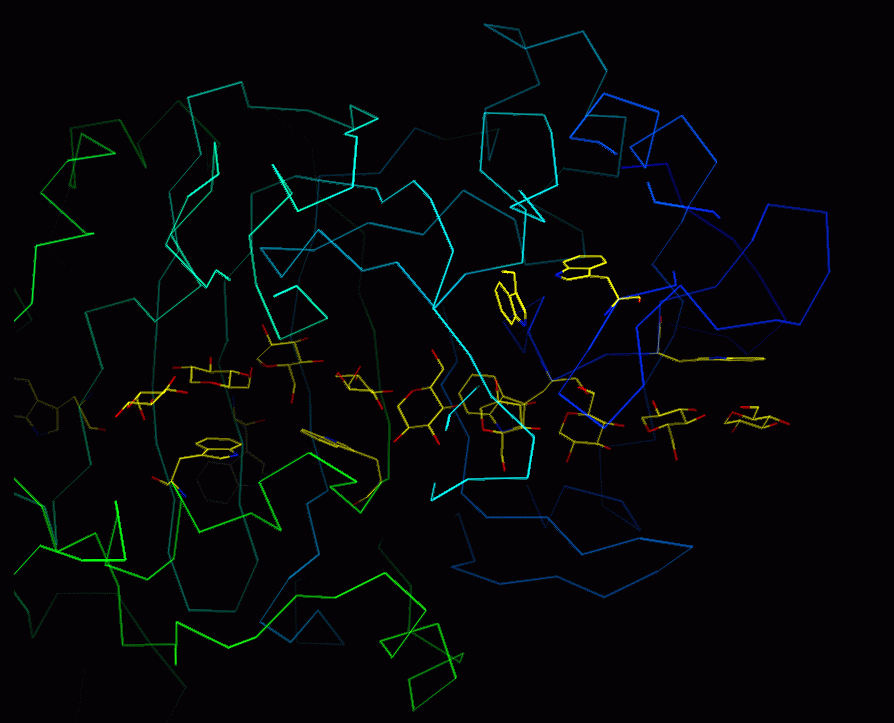

This page describes how to visualise least-squares planes with O and MOLEMAN2. The procedure is illustrated using the structure of cellobiohydrolase I (PDB entry 8CEL, a theoretical model of CBH I in complex with a cellulose nanomer). We will generate planes for each of the nine glucose units, to visualise how the cellulose chain twists its way through the tunnel.
This macro will do the work for one residue at a time. It asks which (glucose) residue number you want to generate a plane for, and the name of the output ODL file. It could loook like this:
----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- ! plane.momac - MOLEMAN2 macro to draw a least-squares plane ! through a glucose residue; only uses the ring atoms to define ! the plane ! ! Gerard Kleywegt @ 990315 ! ! Enter the number of the glucose residue : & resnum ! Enter the ODL file name : & odlfile ! Now select this residue's ring atoms sel none sel or residue GLC sel num and residue_nr $resnum $resnum sel butnot atom " O2 " sel butnot atom " O3 " sel butnot atom " O4 " sel butnot atom " O6 " sel butnot atom " C6 " ! Generate the ODL file for the least-squares plane ono ls_plane $odlfile $resnum yellow 1.0 ! ! All done ! ! ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE -----
As it is, the colour in which the plane will be drawn is hard-wired, as is the margin (1.0 A) around the atoms of the ring for which the plane will be drawn. You can change these values, or use MOLEMAN2 symbols and prompt the user for values (so you could have alternating colours for alternating sugars, etc.).
This we do with MOLEMAN2. First read the molecule (8CEL), then execute the macro we have just written (above):
----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- MOLEMAN2 > read 8cel.pdb Command > (read 8cel.pdb) Reading from file : (8cel.pdb) in normal PDB format ignoring hydrogen atoms ==> Found file in GKPATH : (/nfs/pdb/full/8cel.pdb) HEADER : HYDROLASE 21-NOV-97 8CEL TITLE : THEORETICAL MODEL OF CBH1 IN COMPLEX WITH A CELLULOSE TITLE : 2 NANOMER KEYWDS : HYDROLASE, CELLULOSE DEGRADATION, GLYCOSIDASE, GLYCOPROTEIN, KEYWDS : 2 GLYCOSYLATED PROTEIN EXPDTA : THEORETICAL MODEL AUTHOR : C.DIVNE,J.STAHLBERG,T.A.JONES [...] Nr of selected atoms : ( 3645) MOLEMAN2 > @plane.momac Command > (@plane.momac) ... Opened macro file : (plane.momac) ... On unit : ( 61) Command > (! plane.momac - MOLEMAN2 macro to draw a least-squares plane) Command > (! through a glucose residue; only uses the ring atoms to define) Command > (! the plane) Command > (!) Command > (! Gerard Kleywegt @ 990315) Command > (!) Command > (! Enter the number of the glucose residue :) Command > (& resnum) Value for symbol RESNUM ? ( ) 452 Symbol RESNUM : (452) Command > (! Enter the ODL file name :) Command > (& odlfile) Value for symbol ODLFILE ? ( ) 452.odl Symbol ODLFILE : (452.odl) Command > (! Now select this residue's ring atoms) Command > (sel none) Select NO atoms [...] Eigen value 1 = 12.4 Vector : -0.756840 -0.408495 0.510220 Eigen value 2 = 6.1 Vector : -0.425544 -0.284535 -0.859041 Eigen value 3 = 0.3 Vector : 0.496090 -0.867278 0.041515 Determinant : ( 1.000) Eigenvector #3 defines the least-squares plane Equation: 0.496090 X + -0.867278 Y + 0.041515 Z = -31.307896 ATOM 3240 C1 GLC 452 Distance 0.323 ATOM 3241 C2 GLC 452 Distance -0.192 ATOM 3242 C3 GLC 452 Distance 0.202 ATOM 3243 C4 GLC 452 Distance -0.249 ATOM 3244 C5 GLC 452 Distance 0.212 ATOM 3246 O1 GLC 452 Distance -0.083 ATOM 3250 O5 GLC 452 Distance -0.212 RMSD to plane : ( 0.221) Creating ODL file : (452.odl) Object name : (452) Object colour : (yellow) Border around atoms : ( 1.000) ODL file written Command > (!) Command > (! All done !) Command > (!) ... End of macro file ... Control returned to terminal ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE -----
Simply repeat the procedure for the other eight glucoses (residues 453 to 460), and you're done.
Start up O, and read your molecule (8CEL). Make a CA trace and draw the glucose nonamer. This may look as follows:

Now draw the nine least-squares planes:
----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- draw 452.odl As3> O descriptor in computer file system As3> ODL command ignored: ! draw 453.odl As3> O descriptor in computer file system As3> ODL command ignored: ! [...] dr 460.odl As3> O descriptor in computer file system As3> ODL command ignored: ! ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE ----- EXAMPLE -----
And, hey-presto ! Here's one I prepared earlier:
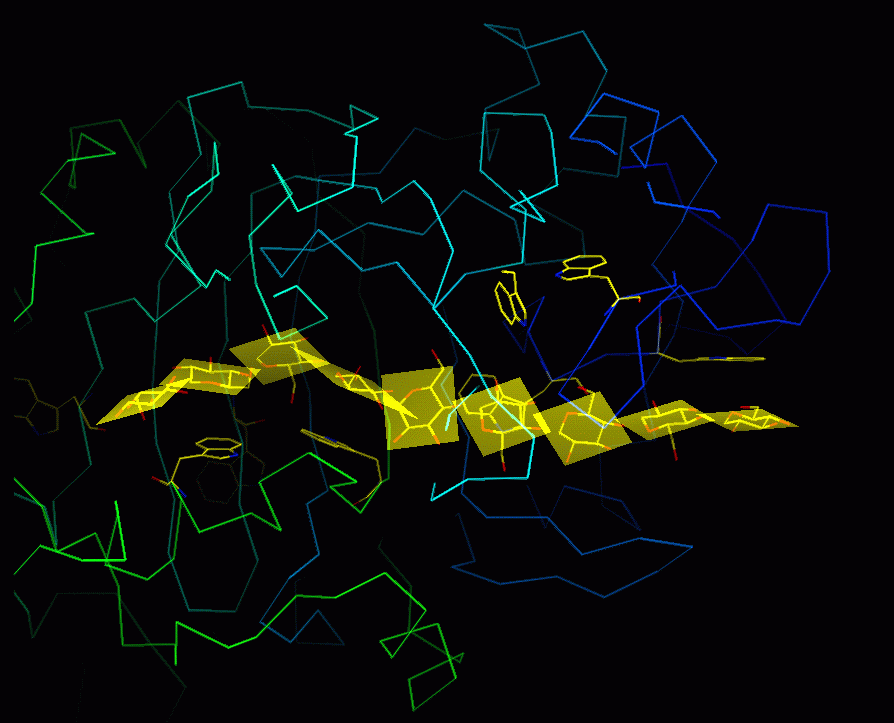
Cute, huh ?
 Latest update at 15 March, 1999.
Latest update at 15 March, 1999.