- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2012
- Structural biology
- Chromodomains in the chloroplast signal recognition particle read a twin-arginine motif
Chromodomains in the chloroplast signal recognition particle read a twin-arginine motif
Chromodomain (CD) containing proteins play important roles in the regulation of gene expression and chromatin remodelling. CDs typically recruit protein complexes to chromatin and read the epigenetic ‘histone code’ by recognising lysine methylation in histone tails. While these canonical CDs all localise to the nucleus, a few are found as cytosolic proteins. The chloroplast signal recognition particle protein 43 (cpSRP43) represents the only known example of CDs implicated in protein targeting.
The signal recognition particle (SRP) is a universally conserved protein-RNA complex that mediates the co-translational targeting of secretory and membrane proteins. Its counterpart in chloroplasts lacks the SRP RNA and is responsible for the post-translational transport of light-harvesting antenna proteins (LHCPs) to the thylakoids [1]. The cpSRP43 protein is a unique addition to the chloroplast SRP (cpSRP) system. It forms a stable complex with cpSRP54 in the stroma and serves as a chaperone for its membrane protein substrate LHCP. Together, these three proteins form the soluble ‘transit complex’ which allows delivery of LHCPs to the thylakoid membrane and subsequent insertion.
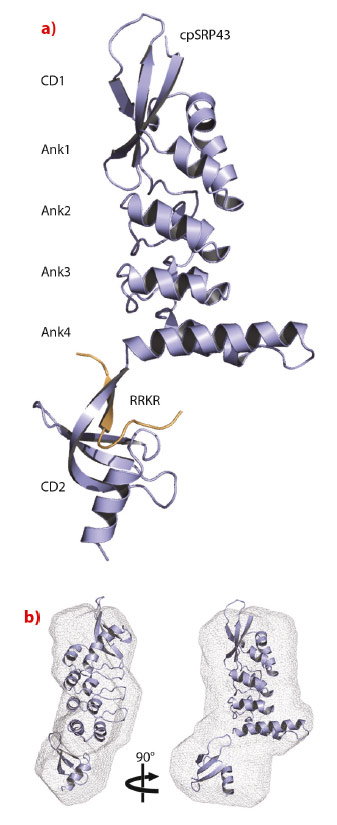
cpSRP43 consists of three chromodomains and four ankyrin repeats [2]. The second chromodomain (CD2) of cpSRP43 binds to the C-terminal tail of cpSRP54, which comprises a conserved arginine-rich motif (RRKR motif). We determined the crystal structure of cpSRP43 from Arabidopsis thaliana in complex with a peptide including the RRKR motif at a resolution of 3.2 Å with data collected remotely at beamline ID14-2 (Figure 17). The structure of the complex in solution was studied by NMR spectroscopy and by small-angle X-ray scattering (SAXS), the latter performed at the BioSAXS beamline ID14-3. CD2 adopts the typical chromodomain fold and the SAXS analysis confirms the overall domain arrangement and indicates flexibility concerning the position of CD2.
 |
|
Fig. 17: Structure of cpSRP43 in complex with the cpSRP54 tail. a) Ribbon presentation of cpSRP43 (ankyrin repeats, Ank1 to 4; chromodomains, CD1 and CD2) and the RRKR peptide in orange. b) Structure of cpSRP43 in solution obtained using SAXS data. The rigid-body model from the crystal structure is superposed within the SAXS ab initio envelope. |
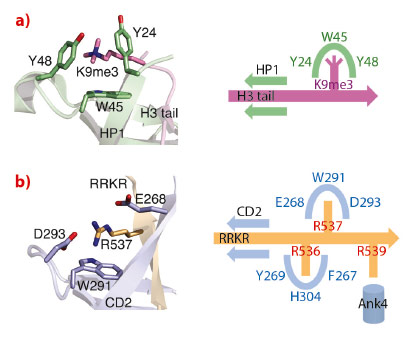
The cpSRP54 tail binds to CD2 via β-completion closing a five-stranded β-barrel. The pattern of β-completion is basically identical to the one observed in canonical, nuclear CDs when binding to histone tails. Methylated lysines in histone tails are accommodated within an aromatic cage formed by three aromatic residues (Figure 18). Cation-π stacking interactions between the positively charged lysine and the electron-rich aromatic ring systems decode the level of methylation (e.g. HP1 recognises the histone H3 tail containing trimethylated lysine 9). The corresponding cage in CD2 is modified and is formed by a single aromatic (Trp291) and two charged residues (Glu268 and Asp293). All three residues are highly conserved and this adaptation of the cage reflects the change in the substrate, with the methylated lysine being replaced by a non-methylated arginine (Arg537). As a unique addition to CD interactions, an adjacent arginine (Arg536) is accommodated in a second aromatic cage formed by three aromatic residues (Phe267, His304 and Tyr269). Therefore, CD2 employs a “twinned cage” to read out a twin-arginine motif using two neighbouring arginines in the cpSRP54 tail. Replacing any of the conserved arginines abolishes complex formation and drastically reduces LHCP insertion.
 |
|
Fig. 18: Substrate recognition by chromodomains. a) An aromatic cage in classical CDs decodes lysine methylation (HP1 with H3K9me3). b) A modified cage in cpSRP43 CD2 reads the second arginine in the RRKR motif (R537). The scheme visualises the twin-arginine motif in the cpSRP54 tail and its read-out by two cages in CD2. |
The evolutionary origin of these non-nuclear CDs is not clear. The earliest occurrences of cpSRP43 in chlorophytes coincide with the appearance of chromovirus retrotransposons in plants. However, strong sequence dissimilarities do not allow for valid conclusions. The transition of chromodomains from reading the epigenetic ‘histone code’ to the recognition of arginines in protein targeting allows employing both chromodomains and the SRP system in a novel functional context. cpSRP43 adapts the universally conserved SRP system to a post-translational function and our structure of cpSRP43 in complex with the C-terminal tail of cpSRP54 demonstrates how the spectrum of chromodomain interactions can be extended.
Principal publication and authors
I. Holdermann (a), N.H. Meyer (b,c), A. Round (d), K. Wild (a), M. Sattler (b,c) and I. Sinning (a), Nature Structural and Molecular Biology 19, 260-263 (2012).
(a) Heidelberg University Biochemistry Center (BZH), Heidelberg (Germany)
(b) Institute of Structural Biology, Helmholtz Zentrum München, Neuherberg (Germany)
(c) Munich Center for Integrated Protein Science and Chair Biomolecular NMR, Technische Universität München, Garching (Germany)
(d) European Molecular Biology Laboratory, Grenoble (France)
References
[1] P. Grudnik, G. Bange and I. Sinning, Biol Chem 390, 775-782 (2009).
[2] K.F. Stengel, I. Holdermann, P. Cain, C. Robinson, K. Wild and I. Sinning, Science 321, 253-256 (2008).



