- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2008
- Structural biology
- Structural biochemistry of a bacterial checkpoint protein
Structural biochemistry of a bacterial checkpoint protein
Maintaining genome integrity is one of the most important processes for the sustainment of life and the prevention of cytotoxic or cancerogenic DNA aberrations. Cells possess various DNA repair pathways as well as DNA damage checkpoint control mechanisms, to ensure that cell cycle progressions occur only with intact chromosomes. In humans, inactivated DNA repair pathways and damage checkpoint controls lead to genetic instability, cancer and ageing syndromes. DisA (DNA integrity scanning A) was recently identified as a checkpoint protein in Bacillus subtilis [1]. It appears to co-localise with sites of DNA double-strand breaks, delaying the cell from entering the sporulation phase. Intriguingly, DisA appears to exist in a single, large complex inside B. subtilis that scans the chromosome for DNA damage. To reveal the molecular and structural mechanism of DisA we used a combination of high-resolution X-ray crystallography, small-angle X-ray scattering (SAXS) and biochemical and biophysical analysis.
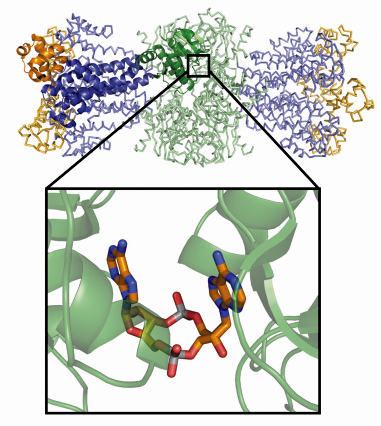
We determined the crystal structure of the Thermotoga maritima DisA homolog (TmaDisA) to 2.1 Å resolution, using multiple-wavelength anomalous dispersion data collected at beamline ID29. According to analytic ultracentrifugation, DisA forms a large octamer with a molecular weight of ~360 kDa. We verified the likely octamer observed in the crystal lattice using SAXS. A protomer of TmaDisA can be divided into 3 domains (Figure 67): an N-terminal globular domain (domain of unknown function, DUF147), a central helical domain and a C-terminal HhH domain. In the octamer eight DUF147 domains form the core of the complex, while the HhH domains form two peripheral tetrameric putative DNA binding platforms.
 |
|
Fig. 67: Overall structure of Thermotoga maritima DisA. One protomer of the octameric assembly is shown in cartoon style: the N-terminal DAC (DUF147)-domain is shown in green, the helical central spine domain in blue and the C-terminal HhH domain in orange. The other chains in the octamer are shown as ribbons. The binding of cyclic-di-adenosine monophosphate at the interface of two DAC-domains is highlighted. |
What is the function of DUF147? Surprisingly, at each of the four interfaces between opposing DUF147 domains we found additional electron density for new types of cyclic dinucleotides. We identified these ligands as cyclic diadenosine monophosphates and have shown that DUF147 of DisA is in fact a diadenylate cyclase. It uses pairs of ATP molecules bound to opposing DUF147 domains for the cyclisation reaction. Thus we suggested renaming the N-terminal domain DUF147 to “DAC-domain” (for diadenylate cyclase). The finding of novel diadenylate cyclase activity and c-di-AMP as product of an enzymatic reaction is quite intriguing, considering the well known and important role of its cousin c-di-GMP as a second messenger in various bacterial processes, including biofilm formation [2]. Possibly, c-di-AMP also acts as messenger, e.g. to coordinate DNA repair and sporulation, or more generally cell division since DisA is also found in non sporulating bacteria.
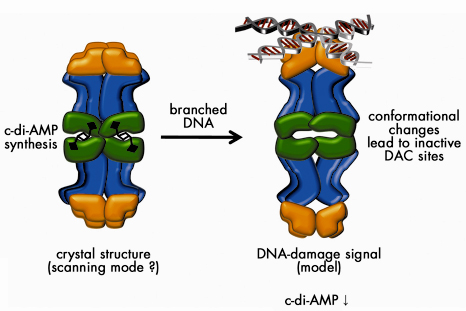
How is di-adenylate cyclase linked to DNA damage scanning? The presence of four HhH-motifs at both ends of the DisA octamer suggests that DisA may not recognise DNA double-strand breaks, but rather its repair intermediates, e.g. Holliday junctions. Gel-electrophoretic mobility shift assays showed that DisA preferentially binds to DNA strands resembling such recombination intermediates but not to plain ssDNA or dsDNA. Consistently, branched DNA substrates, but not plain ssDNA or dsDNA, strongly inhibited the DAC-activity. The structural arrangement of the octamer is ideally suited to allosterically regulate di-adenylate cyclase activity by binding of branched DNA to the HhH-domains array via conformational changes transmitted by the helical middle domain (Figure 68).
 |
|
Fig. 68: Model for the mechanism of action of DisA in checkpoint control. In the absence of chromosomal damage (scanning mode), DisA synthesises c-di-AMP. Recognition of branched nucleic acids such as stalled replication forks or recombination intermediates might inhibit c-di-AMP synthesis thus signalling the presence of unsegregatable chromosomes. Several aspects of the model are speculative, such as the precise mode of DNA binding and the conformational changes shown. |
Our results suggest that DisA may detect sites of unfinished DNA double strand break repair, recombination intermediates or stalled replication forks - all of which should not occur during sporulation or cell division. DisA may signal the presence of such structures via c-di-AMP (or lack of c-di-AMP) although the precise role of c-di-AMP needs to be addressed in future studies. Intriguingly, DUF147 is present in many other bacteria as well as in archaea, arguing for the existence of other c-di-AMP associated processes.
Principal publication and authors
G. Witte (a,b), S. Hartung (a,b), K. Büttner (b,c) andK.-P. Hopfner (a,b), Mol. Cell 30, 167-178 (2008).
(a) Munich-Centre for Advanced Photonics, Munich (Germany)
(b) Center for Integrated Protein Science at the Gene Center, Munich (Germany)
(c) Present address: Max-Planck-Institute for Biochemistry, Martinsried (Germany)
References
[1] M. Bejerano-Sagie et al., Cell 125, 679 (2006).
[2] U. Römling and D. Amikam, Curr Opin Microbiol. 9, 218 (2006).
Acknowledgements
This work was supported by grants from the German Research Council (SFB 684) and the European Union (IP DNA Repair) to K.-P.H.



