- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2004
- Structural Biology
- X-ray Structure of a Minor Group Human Rhinovirus Bound to a Fragment of its Cellular Receptor Protein
X-ray Structure of a Minor Group Human Rhinovirus Bound to a Fragment of its Cellular Receptor Protein
Human rhinoviruses (HRVs) are the main cause of mild upper respiratory disease, usually termed the common cold. The infectious agent is a small icosahedral particle ~300 Å in diameter, composed of a protein shell that encapsidates a single positive RNA strand. The capsid is built from sixty copies, each of four viral proteins VP1, VP2, VP3 and VP4. The first three form the external surface, whereas VP4 is an internal protein in contact with the viral RNA. With more than one hundred immunologically distinct serotypes, HRVs form the largest genus amongst the picornaviridae family. About ninety percent of the HRV serotypes, the major group, use intercellular adhesion molecule 1 (ICAM-1) for cell entry. The remaining serotypes, the minor group, bind to different members of the low-density lipoprotein receptor (LDLR) family. Both receptors are functionally and structurally unrelated. ICAM-1 belongs to the immunoglobulin (Ig) superfamily of proteins whereas the ligand binding domains of LDL receptors are composed of several imperfect direct repeats of about 40 amino acids in length, stabilised by three disulfide bonds and containing a conserved cluster of acidic residues near its C terminal end that coordinates a Ca2+ ion.
The first X-ray structures of intact picornavirus particles, those of a major group rhinovirus HRV14 and of a human poliovirus, revealed a deep cleft or 'canyon' surrounding each pentagonal vertex of the icosahedral shells. This 'canyon' is used by most picornaviruses as an attachment site for binding to their receptors, which typically have Ig-like domains. Three-dimensional electron microscopy image reconstruction techniques provided low resolution images of different rhinovirus-receptor complexes that illustrate this mode of receptor recognition for the major group human rhinoviruses and enteroviruses [1]. In contrast, the receptors for minor group rhinoviruses were shown to bind on a star-shaped dome at the tip of the five-fold vertex [2]. However, high-resolution structures of virus-receptor complexes are required in order to identify the critical interactions that determine the receptor recognition specificity.
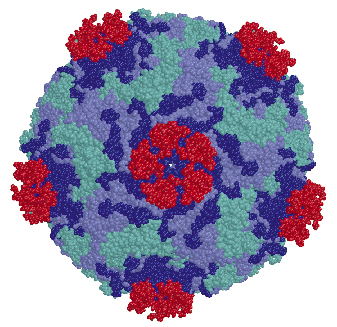 |
Fig. 71: The structure of the HRV2-V3 complex, shown in a space filling representation. Viral subunits VP1, VP2 and VP3 are in blue cyan and lilac respectively and the bound V3 modules are displayed in red. V3 binds outside the "canyon" and close to the virus five-fold vertex. The proximity between the N and C terminal ends of neighbour V3 modules is apparent. |
We determined the co-crystal structure of a minor group human rhinovirus HRV2 with a fragment of one of its receptors, modules 2 and 3 of the very-low density lipoprotein receptor (VLDR). We collected data from three crystals on beamlines ID14-4 and ID29 to a resolution of 3.5 Å and solved the structure by molecular replacement. The averaged maps of the complex showed the presence of only one module of the receptor contacting the viral surface. In this structure the C-terminus of one module is situated very close to the N-terminus of the adjacent module around the five fold axis (Figure 71) suggesting that more than one repeat in a single receptor molecule might attach simultaneously. The binding face of the receptor module is defined by the acidic calcium-chelating residues and, in particular, by an exposed tryptophan that is conserved in a majority of the modules (Figure 72). The attachment site on the virus involves the exposed surface loops BC, DE and HI, derived from two adjacent VP1 subunits, contacting each receptor module at the tip of the fivefold vertex, and one lysine residue in the HI loop, which plays a key roll in receptor recognition. Comparison of the VP1 sequences of all serotypes showed the strict conservation of this lysine in minor group HRVs. The unique combination of hydrophobic and ionic interactions of this lysine seems to be the basis of virus-binding specificity. Nevertheless, other residues in the vicinity must contribute and it is likely that these modulate the affinity of the receptors for the different serotypes.
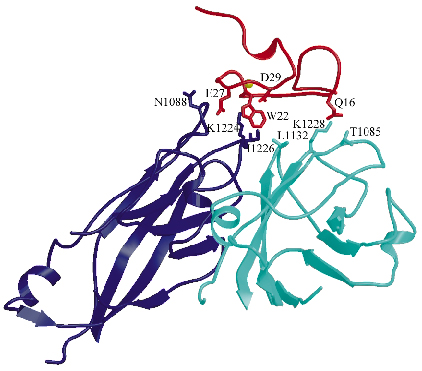 |
Fig. 72: Intermolecular interactions between the V3 module of the receptor (red) and the viral VP1 proteins from two adjacent protomers (dark blue and turquoise, respectively). The Ca2+ ion is represented as a yellow sphere while chains of the residues directly involved in the virus-receptor interactions are shown as sticks. |
References
[1] M.G. Rossmann, Y. He and R.J. Kuhn, TRENDS in microbiology, 10, 324-331 (2002).
[2] E.A. Hewat, E. Neumann, J.F. Conway, R. Moser, B. Ronacher, T.C. Marlovits, and D. Blaas, EMBO J. 19, 6317-6325 (2000).
Principal Publication and Authors
N. Verdaguer (a), I. Fita (a) , M. Reithmayer (b), R. Moser (b), and D. Blaas (b). Nat. Struct. Mol. Biol. 11, 429-434 (2004).
(a) Institut de Biologia Molecular de Barcelona, CSIC Barcelona (Spain)
(b) Vienna Biocenter, University of Vienna (Austria)



