- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2002
- Macromolecular Crystallography
- Structure of the Outer-Membrane Mitochondrial Monoamine Oxidase B
Structure of the Outer-Membrane Mitochondrial Monoamine Oxidase B
Human monoamine oxidases A and B (MAO A and B) are the most intensively investigated flavin-dependent amine oxidases. This is due to their roles in the metabolism of neurotransmitters such as serotonin and dopamine [1,2]. MAO A and MAO B are separate gene products of ~70% sequence identity with both isoforms containing 8 ![]() -S-cysteinyl-FAD coenzymes as the sole redox cofactors and retaining different but partly overlapping substrate and inhibitor specificities. MAOs are implicated in a large number of neurological disorders such as Parkinson's disease and depression and have been important targets for drug therapy over the past 40 years [1]. The three-dimensional structures of both MAO A and MAO B are therefore of interest. However, both enzymes are bound to the outer mitochondrial membrane through a C-terminal polypeptide segment and this feature has made structural investigation by X-ray crystallography more difficult.
-S-cysteinyl-FAD coenzymes as the sole redox cofactors and retaining different but partly overlapping substrate and inhibitor specificities. MAOs are implicated in a large number of neurological disorders such as Parkinson's disease and depression and have been important targets for drug therapy over the past 40 years [1]. The three-dimensional structures of both MAO A and MAO B are therefore of interest. However, both enzymes are bound to the outer mitochondrial membrane through a C-terminal polypeptide segment and this feature has made structural investigation by X-ray crystallography more difficult.
Despite these difficulties, we recently solved the crystal structure of human MAO B using the single isomorphous replacement method combined with multicrystal 12-fold averaging based on two crystal forms grown from different detergent crystallisation conditions. All diffraction data were measured on ESRF beamlines.
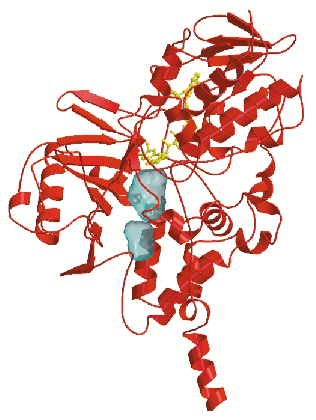
The crystal structure (Figure 1) reveals that MAO B has a two-domain topology similar to that observed in other flavoenzymes. In both crystal forms, the enzyme crystallises as a dimer, which indicates that this oligomeric arrangement probably occurs also in the physiological membrane environment. The 60-residue C-terminal tail (residues 460-520) forms an extended segment that traverses the protein surface and then folds into an ![]() -helix. This helix protrudes from the basal face of the structure with its axis approximately parallel to the molecular two-fold axis, in an orientation suited to anchor the protein to the outer mitochondrial membrane. In addition to this transmembrane helix, two apolar loops located at different positions in the sequence form two hydrophobic patches on the protein surface which are also probably involved in membrane binding. Thus MAO B, which is one of the very few known structures of a monotopically inserted membrane protein appears to have a different method of membrane insertion than those of other monotopically inserted proteins such as prostaglandin synthase and squalene cyclase.
-helix. This helix protrudes from the basal face of the structure with its axis approximately parallel to the molecular two-fold axis, in an orientation suited to anchor the protein to the outer mitochondrial membrane. In addition to this transmembrane helix, two apolar loops located at different positions in the sequence form two hydrophobic patches on the protein surface which are also probably involved in membrane binding. Thus MAO B, which is one of the very few known structures of a monotopically inserted membrane protein appears to have a different method of membrane insertion than those of other monotopically inserted proteins such as prostaglandin synthase and squalene cyclase.
 |
|
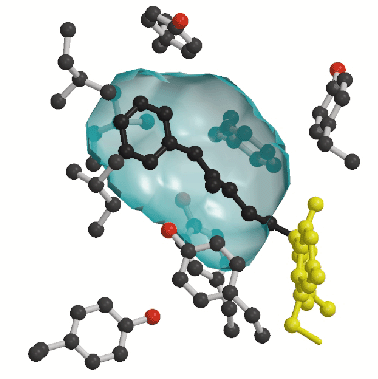
A second prominent feature of MAO B structure is the presence of two adjacent cavities in the interior of the protein (Figure 1). The largest cavity is directly in front of the flavin ring and forms the substrate-binding site occupied by the pargyline inhibitor (Figure 2). For the substrate to enter this cavity, it must first pass through an "entrance cavity" situated near the point where the protein surface intersects with the surface of the outer mitochondrial membrane. This observation raises the intriguing possibility that the anionic membrane surface may function in the electrostatic steering of the positively-charged amine substrate to the entrance cavity binding site. In the substrate cavity, two tyrosyl side chains form an "aromatic sandwich" by facing each other in perpendicular orientations to the flavin, generating an "aromatic cage" that may form the recognition site for the substrate amino group.
 |
|
The features revealed by the three-dimensional structure of MAO B provide a framework for future studies aimed at the investigation of MAO A and MAO B substrate specificities, the functional significance of membrane association and the possible exploitation of the entrance cavity as a site for drug binding.
References
[1] A.M. Cesura, A. Pletscher, Prog. Drug Res. 38, 171-297 (1992).
[2] J.C. Shih, K. Chen, M.J. Ridd, Annu. Rev. Neurosci. 22, 197-217 (1999).
Principal Publication and Authors
C. Binda (a), P. Newton-Vinson (b), F. Hubalek (b), D.E. Edmondson (b), A. Mattevi (a), Nat. Struct. Biol. 9, 22-26 (2002).
(a) Department of Genetics and Microbiology, University of Pavia (Italy)
(b) Department of Biochemistry, Emory University School of Medicine, Atlanta (USA)
 -helix . The figure shows the two cavities found in the substrate-binding domain.
-helix . The figure shows the two cavities found in the substrate-binding domain.


