- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2014
- Structural biology
- A molecular caliper for metered hormone response
A molecular caliper for metered hormone response
Hormones trigger a variety of responses to control growth, development and physiology. Understanding the molecular underpinnings of specificity is a key challenge in biology. The plant signalling molecule auxin is central for growth and development and controls a wide range of processes, including embryogenesis and flower development [1]. Here, we report a molecular mechanism for the generation of diversity and specificity in auxin response.
Auxin is recognised by nuclear receptors, promoting degradation of inhibitor proteins and consequent activation of DNA-binding auxin response factors (ARFs), that control transcription of thousands of genes to bring about a multitude of developmental responses. In the past, we had shown that variations in the ARF family in the higher plant Arabidopsis thaliana largely determine auxin response specificity [2]. For the present research, our biological and structural biology teams joined forces to determine the structural underpinnings of functional diversity within the ARF family.
Crystallographic analysis using beamlines ID14-1 and ID14-4 (now ID30B) revealed that ARF proteins bind DNA as dimers, and let us identify the essential protein-DNA contacts. We then used a range of biochemical and in vivo methods to show that these protein-DNA contacts are indeed essential for DNA binding and function in mediating auxin response and that ARF protein dimerisation is critical for function as it allows high-affinity cooperative binding to complex DNA elements. These elements are composed of inverted repeats of a binding motif with a spacer in between the two motifs.
 |
|
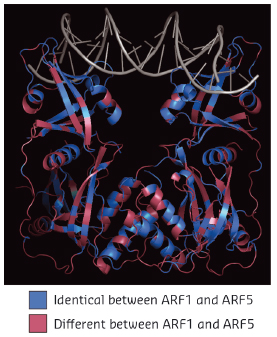
Fig. 118: Structure of Arabidopsis thaliana ARF1 protein dimer in complex with DNA oligonucleotide with two inverted binding motifs. Residues are coloured according to whether they are identical (blue) or different (magenta) between ARF1 and ARF5. |
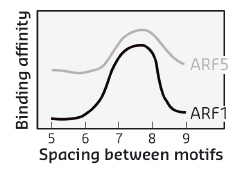
When comparing two highly divergent ARF proteins, we found them to adopt almost identical structures (Figure 118). Likewise, when we determined the DNA-binding preference of ARF monomer proteins, the intrinsic specificity was the same. Thus, neither overall structure, nor intrinsic DNA binding specificity can explain the different biological functions of these two ARF proteins. As the binding of ARF dimers to complex motifs is not only determined by DNA-binding amino acids but also by protein-protein interactions, we tested the hypothesis that ARFs could differ in the spacing tolerated between the two DNA motifs. We found that ARFs differed in this property. While ARF1 could bind with high affinity to complex elements with a spacer of 7 or 8 bases, ARF5 could also bind to motifs with spacing of 5, 6 or 9 bases (Figure 119). Thus, our work reveals a mechanism for DNA-binding specificity within a transcription factor family that is based upon proteins acting as “calipers”. The overall structure of dimers determines the setting of the caliper, and thus the distance between two sites.
 |
|
Fig. 119: Affinity of DNA binding by ARF1 and ARF5 depends on the space between the two inverted binding motifs. |
This work now offers a new framework for understanding specificity in plant hormone response and opens many new avenues for future investigations. Of particular interest would be to discover: how a caliper-based mechanisms is leveraged in living cells to recognise specific genes, how the ARF caliper can be tuned by co-factors and, perhaps most importantly, how this caliper mechanism evolved to increase the complexity of the auxin response.
Principal publication and authors
D.R. Boer (a,b), A. Freire-Rios (c), W.A.M. van den Berg (c), T. Saaki (c), I.W. Manfield (d), S. Kepinski (d), I. Lopez-Vidrieo (e), J.M. Franco-Zorrilla (e), S.C. de Vries (c), R. Solano (e), D. Weijers (c) and M. Coll (a,b), Cell 156, 577-589 (2014).
(a) Institiute for Research in Biomedicine, Barcelona (Spain)
(b) Institut de Biologia Molecular de Barcelona (Spain)
(c) Laboratory of Biochemistry, Wageningen University (Netherlands)
(d) Faculty of Biological Sciences, University of Leeds (UK)
(e) Centro Nacional de Biotecnologia, Madrid (Spain)
References
[1] S. Vanneste and J. Friml, Cell 136, 1005-1016 (2009).
[2] E.H. Rademacher, A.S. Lokerse, A. Schlereth, C.I. Llavata-Peris, M. Bayer, M. Kientz, A. Freire-Rios, J.W. Borst, W. Lukowitz, G. Jürgens and D. Weijers, Dev. Cell 22, 211-222 (2012).



