- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2011
- Structural biology
- Tail-anchored protein targeting to the endoplasmic reticulum membrane
Tail-anchored protein targeting to the endoplasmic reticulum membrane
Eukaryotic cells contain a complex system of membrane surrounded organelles and delivery of newly-synthesised membrane proteins to their specific destinations is of vital importance. Diverse mechanisms for chaperoning membrane proteins have evolved. These include cytosolic factors that identify targeting signals, transiently associate with the hydrophobic regions, and escort their cargo to the designated location.
The best understood targeting machinery is the signal recognition particle (SRP)/Sec61 translocon system at the endoplasmic reticulum (ER) membrane, which accepts membrane proteins with a N-terminal signal [1]. Membrane proteins with a single C-terminal transmembrane helix, the so-called tail-anchored (TA) proteins, utilise the recently identified GET system (guided entry of TA proteins) [2]. The central player in the latter process is the Get3 ATPase, a dimeric protein in which each subunit comprises a nucleotide binding (NBD) and TA binding (TABD) domain. The dimer is held together by a zinc ion and switches between an ‘open’ and a ‘closed’ state by a nucleotide-dependent rotation of the two subunits. At the ER membrane, Get3 interacts with the cytoplasmic domains of the two receptor proteins Get1 and Get2, which are necessary and sufficient for TA protein insertion [3]. To dissect the mechanism of TA protein insertion we studied the structure and function of the Get3 ATPase in complex with its receptors Get1 and Get2.
Due to the inherent flexibility of a molecular targeting machine, crystal growth proved difficult and the success of the project relied on the brilliant beamlines ID14-4, ID23-1 and the microfocus ID23-2 coupled with regular access via the BAG scheme.
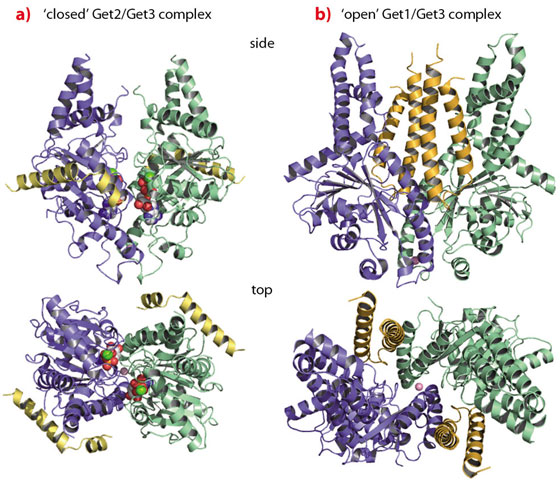
The crystal structure of Get3 in complex with the cytoplasmic domain of Get1 (Get1 for simplicity), determined at a resolution of 3.0 Å, revealed Get1 to form a typical coiled-coil stabilised by a hydrophobic zipper in a heptad repeat. The Get1/Get3 complex forms a symmetrical heterotetramer with a central Get3 homodimer and two Get1 molecules wedging into the Get3 dimer interface (Figure 104b). Get1 stabilises nucleotide-free ‘open’ conformations of Get3 by directly interfering with nucleotide binding and rendering the active site highly solvent accessible. Each Get1 molecule binds to both Get3 subunits, however, the interactions are different in size and strength. While the larger interface performs a structural role and seems independent of Get3 closure, the smaller interface ‘slides’ along the second Get3 subunit during opening to finally convey nucleotide exchange activity in the ‘open’ conformation.
 |
|
Fig. 104: TA protein targeting by the GET system. a) The ‘closed’ Get2/Get3 complex; b) the ‘open’ Get1/Get3 complex. |
Since both Get1 and Get2 are necessary for TA protein insertion, we solved the structure of the Get2 N-terminal cytoplasmic domain in complex with Get3 at 4.6 Å resolution. The Get2/Get3 complex is also a symmetric heterotetramer with two Get2 molecules bound to the nucleotide-loaded ‘closed’ Get3 homodimer (Figure 104a). The Get2 interaction region comprises two amphipathic, positively charged helices connected by a flexible glycine linker. Each Get2 molecule interacts laterally with a negatively charged surface patch of a single Get3 subunit, and does not interfere with nucleotide binding. Interestingly, this binding site partially overlaps with the binding site described for Get1 as also confirmed by NMR titration experiments. Thus, sequential binding of Get2 and Get1 to the Get3/TA protein complex might be envisaged, which is accompanied by structural rearrangements in the targeting complex at the membrane.
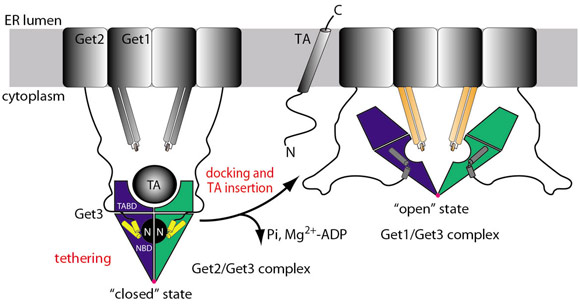
The integration of the crystal structures with biochemical data suggests the following mechanistic model for TA protein delivery to the ER (Figure 105): The Get3/TA protein complex is tethered to the membrane by the N-terminus of Get2. Docking of Get1 to the tethering complex partially displaces Get2, which results in an insertion competent complex. Insertion of the TA protein allows for Pi release and the stored energy of ATP hydrolysis drives Get3 from the ‘closed’ to the ‘open’ state as observed in the Get1/Get3 structure with a concomitant loss of Mg2+-ADP. As the cytoplasmic domain of Get1 is rigidly linked to its transmembrane regions, the rearrangements during opening of Get3 may create a force, which could be directly exploited for TA protein insertion by a mechanism reminiscent of ABC transporters. Finally, Get3 dissociates from the membrane by ATP binding and adopts the ‘closed’ conformation, ready for the targeting of the next TA protein.
 |
|
Fig. 105: Mechanistic model of TA protein targeting by the GET system. Coloured parts correspond to structures shown in Figure 104: Get3 dimer (blue/green), Get2 (yellow), and Get1 (orange). |
Principal publication and authors
S. Stefer (a), S. Reitz (b), F. Wang (c), K. Wild (b), Y.-Y. Pang (b), D. Schwarz (d), J. Bomke (d), C. Hein (a), F. Löhr (a), F. Bernhard (a), V. Denic (c), V. Dötsch (a) and I. Sinning (b), Science 333, 758-762 (2011).
(a) Institute for Biophysical Chemistry, Centre for Biomolecular Magnetic Resonance, Goethe University, Frankfurt am Main (Germany)
(b) Heidelberg University Biochemistry Center (BZH), Heidelberg (Germany)
(c) Department of Molecular and Cellular Biology, Harvard University, Northwest Laboratories, Cambridge (USA)
(d) Merck KGaA, Merck Serono, Molecular Interactions and Biophysics, Darmstadt (Germany)
References
[1] P. Grudnik, G. Bange and I. Sinning, Biol Chem 390, 775-782 (2009).
[2] P.J. Simpson, B. Schwappach, H.G. Dohlman and R.L. Isaacson, Structure 18, 897-902 (2010).
[3] F. Wang, A. Whynot, M. Tung and V. Denic, Mol Cell 43, 738-750 (2011).



