- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2010
- Structural biology
- Repairing breaks in DNA double strands: the structure of DNA-PKcs
Repairing breaks in DNA double strands: the structure of DNA-PKcs
Breaks in both strands of chromosomal DNA occur frequently and constitute a major threat for cells. Left unrepaired or mis-repaired they can result in cell death or cancer. In eukaryotes, two different pathways are used to repair this type of DNA damage, the balance between them varying during the cell cycle [1]. Unlike homologous recombination, which requires a sister chromatid, non-homologous end joining [2] does not copy information lost in double-strand breaks and so can be active throughout the cell cycle. In this process two DNA ends are joined directly, with the help of many proteins that organise, tailor and ligate the broken ends. An important component of this process is DNA-dependent protein kinase (DNA-PK), an enzyme that comprises the DNA-PK catalytic subunit (DNA-PKcs) and the Ku70/Ku80 heterodimer. DNA-PKcs is a protein comprising a single polypeptide chain of 4128 amino acids [3] that belongs to a family of enzymes involved in the sensing and transmission of DNA damage signals initiating cell cycle arrest. Knowledge of its architecture should lead to a better understanding of its role in the events that take place in joining DNA ends. Although electron micrographs have provided images of this large enzyme, crystals have previously been elusive.
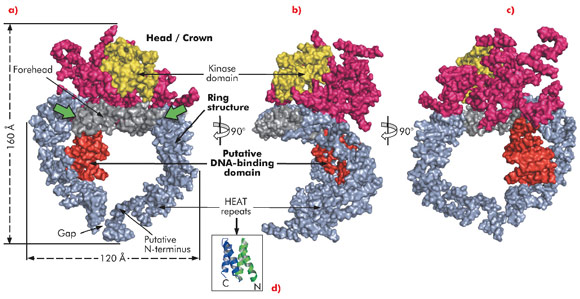
Here, we describe the crystal structure of DNA-PKcs in complex with the C-terminal domain of Ku80 at 6.6 Å resolution (Figure 115) in which the overall fold of this enzyme is clearly visible for the first time. The structure was solved using the multi-wavelength anomalous dispersion (MAD) method exploiting a Ta6 Br122+ heavy metal cluster introduced into the crystals and using diffraction data collected on beamline ID29. At 6.6 Å resolution the electron density of helical regions is clearly visible indicating that the structure is dominated by α-helices arranged as HEAT repeats. However, the electron density for the loops that link these helices is not visible thus making it difficult to fit the whole molecule reliably. Furthermore, as the Ku80 C-terminal domain also consists of α-helical HEAT repeats it has not yet been possible to locate this moiety in the electron density.
The long polypeptide chain of DNA-PKcs is organised into several distinct domains (Figure 115). From the N-terminus 66 helices are arranged as HEAT repeats and folded into a hollow circular structure, which has a concave shape. Just before the circle is complete, the polypeptide chain changes direction, leaving a gap, and folds into a much smaller globular domain also organised as HEAT repeats that sits on one side of the circular structure (red in Figure 115a). DNA-PKcs bind directly to DNA [4] and this domain is a good candidate for being the DNA binding domain. After this domain the polypeptide chain then crosses the circular structure and forms the larger C-terminal head/crown domain that is perched on top of the circular structure. This region is also predominantly α-helical and is likely to interact with many other proteins. Contained in this region is the protein kinase domain, involved in both autophosphorylation and the phosphorylation of other proteins.
In summary, the ring structure reflects the role of DNA-PKcs both as an enzyme involved in DNA damage signalling and as a platform for the binding of DNA, Ku and other proteins engaged in the repair of broken DNA. The head/crown kinase domain located at the very top of the circular structure is well placed for easy access to substrates that are phosphorylated by this protein. There are some irregular regions in the arrangement of the HEAT repeats within the ring structure (Figure 115a, green arrows). These may accommodate conformational changes that widen the gap, shown in Figure 115a, thus providing a way for the molecule to come off DNA at the end of the repair process.
Principal publication and authors
B.L. Sibanda, D.Y. Chirgadze and T.L. Blundell, Nature 463, 118–121 (2010).
Department of Biochemistry, University of Cambridge (UK)
References
[1] M. Shrivastav, L.P. De Haro and J.A. Nickoloff, Cell Research 18, 93-113 (2008).
[2] S. Critchlow and S.P. Jackson, Trends Biochem. Sci. 23, 394-398 (1998).
[3] K.O. Hartley, D. Gell, G.C. Smith, H. Zhang, N. Divecha, M.A. Connelly, A. Admon, S.P. Lees-Miller, C.W. Anderson and S.P. Jackson, Cell. 82, 849-856 (1995).
[4] M. Yaneva, T. Kowalewski and M.R. Lieber, EMBO J. 16, 5098-5112 (1997).