- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2008
- X-ray imaging and optics
- Stem cell characterisation in the human intestine derived by mid-infrared light
Stem cell characterisation in the human intestine derived by mid-infrared light
The human body is comprised of different cell types, which together form complex tissues. Cells are evolved structures composed of biomolecules, such as proteins, lipids, DNA, RNA, glycoproteins and carbohydrates; their make-up differs with function. Stem cell populations are believed to be slow-cycling, possessing an infinite proliferative capacity and giving rise to transit-amplifying (TA) cells; these then differentiate into functional terminally-differentiated (TD) cells.
Identification of stem cells is important in order to understand fundamental tissue biology. However, robust markers for adult stem cells remain elusive because many approaches have relied on the expression of a single cell-surface marker. The most regenerative tissue in the human and an important stem-cell system which should be amenable to characterisation, due to its well-compartmentalised structure, is the gastro-intestinal (GI) tract containing the crypts of the small and large intestine. The GI tract is a model system as the putative stem cells are generally believed to reside towards the base of the crypt; these divide slowly migrating upwards to form TA cells, which form TD cells after further cell divisions. It has always been something of a conundrum that we seem to know a great deal about intestinal stem cells and yet reliable markers have proved to be elusive. However, it is generally agreed that in the small intestinal crypt, the stem cells are located, at approximately cell positions 4-5 (counting from the crypt base), whereas in the large intestinal crypts, they are at the very base.
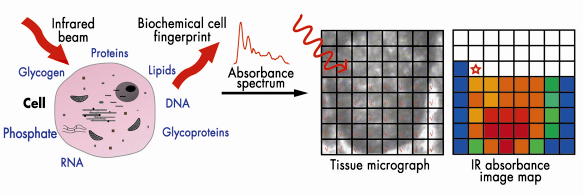
Because of their different in situ roles, one might expect that these cell populations would have differing biochemical profiles giving differing infrared spectral fingerprints (Figure 135). Cellular biomolecules absorb the mid-infrared (![]() = 2-20 µm) via vibrational transitions that are derived from individual chemical bonds; this may yield richly structured ‘‘fingerprint’’ spectra relating to structure and conformation [1]. Synchrotron infrared sources such as at ESRF generate a highly collimated beam of photons of high brilliance that may be delivered through a very small aperture.
= 2-20 µm) via vibrational transitions that are derived from individual chemical bonds; this may yield richly structured ‘‘fingerprint’’ spectra relating to structure and conformation [1]. Synchrotron infrared sources such as at ESRF generate a highly collimated beam of photons of high brilliance that may be delivered through a very small aperture.
 |
|
Fig. 135: Biomolecules absorb in the mid-infrared allowing interrogation of cells with differing biochemistry. Chemical entities may be tracked in a pixel-by-pixel fashion. |
We set out to determine whether specific biomolecular fingerprints derived using infrared microspectroscopy could be attributed to different regions. Marked differences (including peak location and intensity) were observed throughout the infrared spectral “fingerprint” region (900 cm–1 to 1,800 cm–1). Large numbers of variables sometimes make it difficult to identify underlying variance [2]. Such data may be interrogated using principal component analysis (PCA). PCA was used for data reduction, and the output was processed using linear discriminant analysis (LDA). This allowed new variables to be found such that the ratio of the between-cluster (i.e., category of cell type based on locations) variance to the within-cluster variance was maximised. PCA-LDA highlighted ![]() sPO2– (1,080 cm–1) as being a major contributor to segregation, pointing to an important role of alterations in the structure of chromatin.
sPO2– (1,080 cm–1) as being a major contributor to segregation, pointing to an important role of alterations in the structure of chromatin.
 |
|
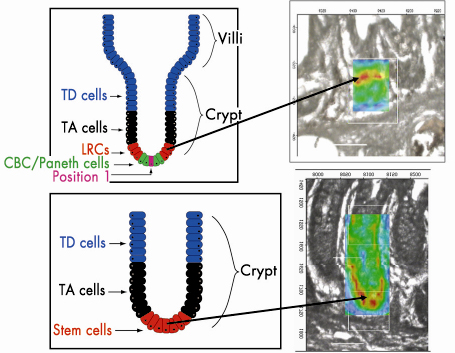
Fig. 136: Stem cell hierarchy of small and large intestine stem cells. Infrared spectral image maps highlight |
Infrared spectral image maps allow one to track the spatial distribution of chemical entities based on levels of relative absorbance intensity at a wavenumber in a pixel-by-pixel fashion (Figure 136). Tissues analysed at the high resolution (8 µm x 8 µm) possible at ESRF generated such image maps that were then superimposed exactly on the interrogated regions (Figure 136). Our findings highlighted ![]() sPO2– with greatest intensity located in a ring in the small intestinal crypt, at approximately cell positions 4-5 near the base, whereas in the large bowel crypts, this occurred primarily at cell positions 1-4, at the base. This suggests that this approach is a powerful adjunct to our understanding of stem cell biology.
sPO2– with greatest intensity located in a ring in the small intestinal crypt, at approximately cell positions 4-5 near the base, whereas in the large bowel crypts, this occurred primarily at cell positions 1-4, at the base. This suggests that this approach is a powerful adjunct to our understanding of stem cell biology.
Principal publication and authors
M.J. Walsh (a), T.G. Fellous (b), A. Hammiche (a), W.R. Lin (b), N.J. Fullwood (a), O. Grude (a), F. Bahrami (c), J.M. Nicholson (c), M. Cotte (d), J. Susini (d), H.M. Pollock (a), M. Brittan (b), P.L. Martin-Hirsch (a), M.R. Alison (b) and F.L. Martin (a), Stem Cells 26, 108 (2008).
(a) Lancaster University (UK)
(b) Queen Mary’s School of Medicine and Dentistry (UK)
(c) Daresbury Laboratory (UK)
(d) ESRF
References
[1] M.J. Walsh, M.J. German, M. Singh, H.M. Pollock, A. Hammiche, M. Kyrgiou, H.F. Stringfellow, E. Paraskevaidis, P.L. Martin-Hirsch and F.L. Martin, Cancer Lett. 246, 1 (2007).
[2] M.J. German, A. Hammiche, N. Ragavan, M.J. Tobin, N.J. Fullwood, S.S. Matanhelia, A.C. Hindley, C.M. Nicholson, H.M. Pollock and F.L. Martin, Biophys. J. 90, 3783 (2006).



