- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2007
- Structural Biology
- Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit
Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit
The trimeric influenza virus polymerase, comprising subunits PA, PB1 and PB2, is responsible for transcription and replication of the segmented viral RNA genome [1]. It is of significant interest because structural characterisation may permit design of a new class of anti influenza drugs, and because mutations of the subunits can lead to avian-to-human infection. Despite intensive work, structural studies have been prevented by an inability to express the proteins, either individually or as a complex.
 |
|
Fig. 69: Identification of PB2 C-terminal domain via a colony-based expression screen of 26,880 random deletion constructs of the pb2 gene. Stable expression of soluble protein results in efficient in vivo labelling of a C-terminal biotin acceptor peptide. Detection is by fluorescent streptavidin and fluorimager. |
 |
|
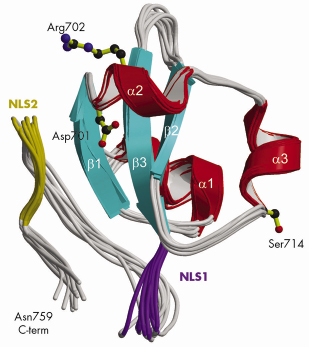
Fig. 70: Ribbon diagram of the 10 lowest energy NMR structures of the C-terminal PB2 domain showing Asp701, Arg702 and Ser714, which are implicated in cross-species transmission, and basic regions corresponding to the minor (purple) and major (gold) sites of the bipartite NLS. |
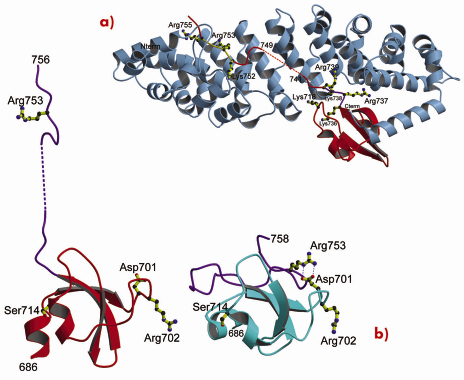
Using a novel random library screening technique for “difficult-to-express” proteins called ESPRIT (Expression of Soluble Proteins by Random Incremental Truncation), developed at the EMBL, we screened 27,000 genetic constructs to identify an independently folded C-terminal PB2 domain (Figure 69). The structure was initially solved by NMR at the IBS (Figure 70) revealing a well-packed domain containing three surface residues implicated in adaptation from avian to mammalian hosts. Analysis of the sequence of the flexible C-terminus suggested the presence of a previously overlooked bipartite nuclear localisation sequence (NLS). This putative function was confirmed using GFP (green fluorescent protein) fusions; we showed that both the domain and full-length PB2 subunit are efficiently imported into the nucleus dependent on the NLS (not shown). The 2.2 Å crystal structure of the domain complexed with human nuclear import factor importin a5 (data collected on beamline ID23-1) revealed how the last twenty residues of the domain unfold to permit binding within the superhelical groove of the importin in a classical manner (Figure 71a). A comparison of the two structures (Figure 71b) shows that one of the cross-species mutation residues (D701) tethers the NLS-containing peptide to the core of the domain in the unbound state via a salt bridge with Arg753. This interaction is broken when the NLS is unfurled during importin binding and suggests that the mutation D701N, observed in some pathogenic strains of influenza including H5N1 infecting humans in Vietnam [2], may alter the energetics of this interaction.
 |
|
Fig. 71: X-ray structure of the PB2 C-terminal domain complexed with importin |
In conclusion, by exhaustive random construct screening, we identified an unsuspected domain at the C-terminus of the PB2 subunit and determined both the NMR solution structure of the isolated domain and the X-ray structure of a complex with the nuclear import receptor importin a5. Cell biology studies confirmed the role of the domain in nuclear localisation of the polymerase subunit. A comparison of the two structures sheds light on a new mechanism of avian-to-human transfer of influenza viruses, including some current H5N1 strains.
References
[1] D.L. Noah and R.M. Krug, Adv. Virus Res. 65 121-45 (2005).
[2] M.D. de Jong, et al. Nat Med 12, 1203-7 (2006).
Principal publication and authors
F. Tarendeau (a), J. Boudet (b), D. Guilligay (a), P. Mas (a), C. Bougault (b), S. Boulo (c), F. Baudin (c), R.W.H. Ruigrok (c), N. Daigle (d), J. Ellenberg (d), S. Cusack (a), J.-P. Simorre (b) and D.J. Hart (a) Nat Struct Mol Biol. 14 229-33 (2007).
(a) EMBL, Grenoble (France)
(b) IBS, Grenoble (France)
(c) UVHCI, Grenoble (France)
(d) EMBL, Heidelberg (Germany)



