- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2007
- Structural Biology
- Structural insights on synthesis of inflammatory mediators by human leukotriene C4 synthase
Structural insights on synthesis of inflammatory mediators by human leukotriene C4 synthase
Leukotriene C4 synthase is an integral membrane protein that catalyses the conjugation between the reactive epoxide on the lipidic leukotriene A4 (LTA4) and the cysteinyl sulfur on glutathione (GSH). The product, LTC4, together with its breakdown products (LTD4 and LTE4) are referred to as cysteinyl leukotrienes (Cys-LTs) or the “slow reacting substance of anaphylaxis”. These molecules are powerful mediators of inflammation and cause constriction of smooth muscle cells in the airways as well as myocardial contractility. Drugs targeting the signalling pathways of Cys-LTs have offered therapeutic opportunities in the medical treatment of asthma.
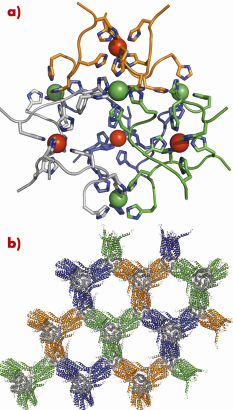
The structure of human LTC4S was solved using the MAD phasing technique using a Pt derivative, which, together with 2.0 Å data on the apo protein, was collected on ID14-4. We also solved a GSH complexed structure of LTC4S to 2.2 Å using data from ID23-2. The use of the highly-automated stations with sample changers and a robust setup, allowed us to screen through more than 1000 crystals in the search for bound heavy atoms and ligands as well as well-diffracting crystals. The crystals grew in a cubic space group, assigned as the highly unusual F23, and contained only ~20% protein. The fact that the crystals diffract so well despite the high solvent content is interesting and is possibly due to the high symmetry space group, as well as an intriguing 6-His polynuclear metal cluster that effectively ordered the crystals (Figure 80a and 80b).
 |
|
Fig. 80: a) Metal cluster composed of 12 crystallographically related hexa-histidine tags from four trimers (colored respectively in green, orange, blue and light grey). Each trimer coordinates one metal internally (shown as a red sphere) and one monomer from three different trimers coordinates another metal (shown as a green sphere). In total, 48 histidine residues coordinate 8 metals. (b) Crystal packing, trimers are coloured green, orange, grey or blue. |
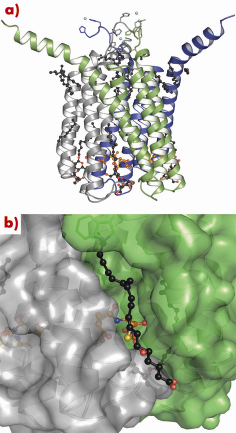
LTC4S is a compact homo-trimeric protein (Figure 81a) with a total of 12 membrane-spanning helices and three active sites buried in deep polar pockets at the interface of two monomers. A relatively long loop that connects helices 1 and 2 folds on top of the neighbouring monomer and contributes to subunit interactions. This loop also seals off the cytosolic entrance to the active site and is proposed to have some flexibility to allow GSH to enter. The active site needs to be accessible both to the lipidic substrate LTA4, which may diffuse through the membrane layer, and to the highly water soluble co-substrate GSH, which can access from the cytosol. The active site forces the reactive cysteinyl thiolate on GSH towards the protein/membrane interface, where LTA4 binds in a hydrophobic crevice formed between two monomers (Figure 81b). This open crevice stretches from the cytosolic face of the protein towards the peri-nuclear side where the most distal part is covered by a tryptophane side chain that serves as a lid on the crevice. In our electron-density maps we also see a number of extended features, likely to be due to lipids or detergents from the purification procedure that line the protein surface. Several of these can be found in, or near, the LTA4 binding crevice and serve as a good substrate model because they have important structural similarities to LTA4.
 |
|
Fig. 81: (a) Side view of a trimer, monomers coloured grey, blue and green. Lipid and detergent molecules are visualised as black balls and sticks, the GSH backbone as orange balls and sticks. Metal ions are shown as cyan spheres. (b) Close-up, transparent surface view of (a) showing the binding crevice for docked LTA4. The GSH that contains the thiolate is shown in yellow. |
The different binding modes of aliphatic chains in the presence or absence of GSH give us a clue as to how recognition of LTA4 and the positioning of the reactive epoxide is achieved. We propose that GSH binding precedes that of LTA4 and assists in the formation of an appropriate lipid-binding crevice for LTA4.
In conclusion, our structure shows how a fairly small protein can accommodate and recognise substrates with widely different chemical properties; the enzyme buries a highly-soluble substrate and presents it to a lipid for conjugation, a reaction that would never occur unless aided. These structural insights into the mechanism of LTC4 formation could lead the way for development of new drugs in the treatment of asthma and chronic inflammation.
Principal publication and authors
D. Martinez Molina (a,b), A. Wetterholm (a), A. Kohl (a), A.A. McCarthy (c), D. Niegowski (a,b), E. Ohlson (a), T. Hammarberg (a), S. Eshaghi (a), J.Z. Haeggstrom (a), P. Nordlund (a), Nature 448, 613 - 616 (2007).
(a) Karolinska Institutet (Sweden)
(b) Stockholm University (Sweden)
(c) EMBL Grenoble Outstation (France)



