- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2007
- Structural Biology
- Structural insights into the early steps of homologous recombination in bacteria
Structural insights into the early steps of homologous recombination in bacteria
DNA damage is a common occurrence that compromises the functional integrity of our genome. Well over 10,000 instances of DNA damage are estimated to occur daily in every human cell. The causative agents of the damage are mainly free radicals that are produced as natural by-products of food metabolism. If damaged DNA is left unrepaired, it generates mutations, replication errors, persistent DNA damage and genomic instability, which are ultimately associated with cancer and ageing. The mechanisms of DNA repair at a molecular level are largely unknown and a better understanding of the detailed mechanisms and principles underlying damage recognition in prokaryotes is an essential step towards obtaining a complete overview of the more complex human DNA repair systems. In prokaryotes, four repair pathways exist (homologous recombination, base-excision repair, nucleotide-excision repair and mismatch repair), all of which are essential for viability.
Homologous recombination, in addition to its fundamental role in genetic diversification of bacterial genomes, plays a key role in the repair of a variety of DNA lesions, including the lethal double-strand breaks. Initiation of homologous recombination can be carried out by either the RecBCD or the RecFOR proteins; in both cases these proteins act as mediators for RecA binding to single-stranded DNA in order to allow for homologous strand invasion. Genome sequence analysis of Deinococcus radiodurans, an outstanding bacterium capable of repairing and recovering from several hundred DNA double-strand breaks in its genome, has revealed that the genes encoding RecB and RecC proteins are missing, while all the genes encoding members of the RecFOR pathway (RecQ, RecJ, RecO, RecF and RecR) are present [1]. Inhibition of the RecFOR pathway in D. radiodurans increases the sensitivity of D. radiodurans cells to gamma-radiation, suggesting that RecFOR is essential for the repair of double-strand breaks.
 |
|
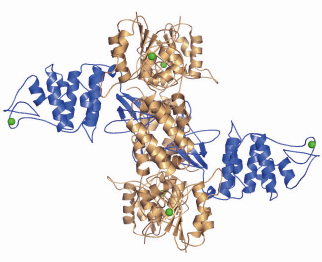
Fig. 74: Crystal structure of the hetero-hexameric complex formed between RecO (blue) and RecR (bronze). Zn2+ ions are illustrated as green spheres. |
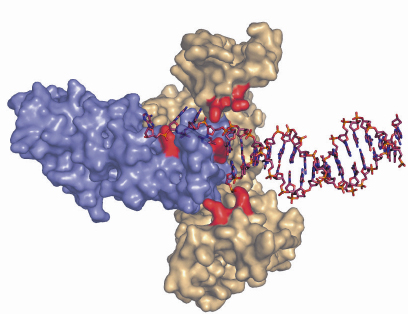
An increasing amount of structural information is becoming available for these proteins, which together with the biochemical and genetic data will no doubt improve our current understanding of the initial steps in homologous recombination. The individual crystal structures of RecF, RecO [2] and RecR from D. radiodurans have been solved over the past three years allowing the mapping of regions involved in protein-protein and protein-DNA interactions. To gain a better understanding of the detailed mechanisms involved in RecFOR-mediated recombination events, we have determined the crystal structure of the RecOR complex from D. radiodurans at 3.8 Å resolution using data collected at beamline ID14-2. The complex is a hetero-hexamer comprising two RecO molecules and four RecR molecules in which the RecO molecules are positioned on either side of the tetrameric ring of RecR, obstructing access to the interior of the ring (Figure 74). The interactions between RecO and RecR observed in our low resolution crystal structure were confirmed by mutagenesis and binding studies. This work identified a key ionic interaction between RecO and RecR as being essential for the stability of the complex. Furthermore, the structural data served as a framework for designing mutants to investigate the ability of RecOR to bind DNA. Similar studies had previously been carried out using only the individual RecO and RecR proteins. This study revealed that two regions of the complex, one on RecO and another on RecR are critical for DNA binding. Both of these regions are located in the centre of the complex and would not be accessible for DNA binding in the configuration of RecOR observed in our crystal structure. Our mutagenesis and biochemical analyses taken together with the three-dimensional arrangement of the RecOR hetero-hexameric complex thus strongly indicate that a rearrangement of the complex needs to take place in order to allow DNA binding to occur. Chemical cross-linking studies indicate that RecOR most likely undergoes both local conformational changes and a larger architectural reorganisation upon binding to sites of double-strand break repair (Figure 75). Displacement of one of the two RecO molecules seems to be a prerequisite for DNA binding, but disruption of the tetrameric ring of RecR may also be needed.
 |
|
Fig. 75: Hypothesis of the binding of a structurally-rearranged hetero-trimeric complex of RecOR to double-stranded DNA with an extended single-stranded 3’overhang. Residues identified as being critical for DNA binding are coloured in red. |
This study has shed light on the importance of studying protein complexes in addition to the individual proteins. Many of the features described for the single proteins were no longer observed within the context of the complex. Protein-protein and protein-DNA interactions are key to regulating many cellular processes. A better understanding of the processes regulating complex formation between the various Rec partners will certainly help in establishing the detailed sequence of events leading to RecA-dependent homologous recombination.
References
[1] O. White, et al., Science, 286, 1571-1577(1999).
[2] I. Leiros, J. Timmins, D.R. Hall, S. McSweeney, EMBO J., 24, 906-918 (2005).
Principal publication and authors
J. Timmins (a), I. Leiros (b) and S. McSweeney (a). EMBO J., 26, 3260-3271 (2007).
(a) ESRF
(b) The Norwegian Structural Biology Centre (NorStruct), University of Tromsø (Norway)



