- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2005
- Soft Condensed Matter
- Structural Basis of the Regulation of Cell Adhesion in Brain Development and Disease
Structural Basis of the Regulation of Cell Adhesion in Brain Development and Disease
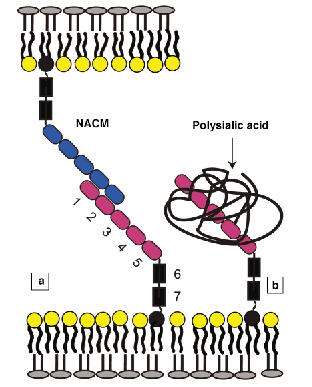
The neural cell adhesion molecule (NCAM) is one of the most abundant adhesion proteins in the central nervous system. It is critical in brain development, directing cell growth and forming adhesive contacts between neural cells. The polypeptide chain folds into seven linearly arranged 3.5-4 nm domains (Figure 62a). NCAM mediates intercellular adhesion by binding to identical proteins on adjacent cells (Figure 62a). One of the significant features of NCAM is that it occurs in both adhesive and anti-adhesive forms. The anti-adhesive form contains 1-2 linear polymers of sialic acid (PSA) attached to the fourth domain (Figure 62b). This modification dramatically affects both the architecture of neural tissues and biological function [1]. PSA-NCAM increases the spacing between cells in tissues, it increases neural plasticity, allowing cells to more easily break and form contacts, and the abnormal expression of PSA-NCAM in cancer increases tumour aggressiveness.
 |
|
Fig. 62: (a) Overall structure of NCAM on the cell membrane. The seven-domain protein binds to an identical NCAM on the opposing membrane to form cell-cell junctions. (b) PSA-NCAM is modified with up to two linear polysialic acid chains, which are negatively charged. |
One model for the molecular basis of PSA’s biological activity postulates that PSA increases the steric repulsion between cells, and counteracts protein binders [1]. This requires that the excluded volume of PSA be sufficient to prevent the approach of adhesion proteins on adjacent cells. X-ray reflectivity studies at the Troika beamline at ID10B quantified the effect of PSA on the hydrodynamic volume of NCAM, and tested the hypothesis that the excluded volume of the PSA acts as a steric modulator of NCAM’s adhesive function.
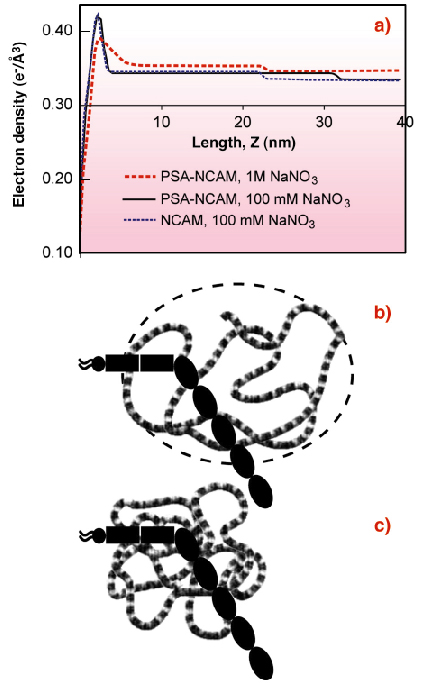
The electron density profile of an oriented NCAM monolayer immobilised on a lipid monolayer at the liquid-vapour interface of a Langmuir trough (Figure 63a) revealed that the protein is bent or tilted at the membrane (Figure 63b). The end-to-end length of the NCAM extracellular region is 27 nm, but the protein box thickness was only 20.6 nm. Neutron reflectivity profiles of NCAM bound to a supported bilayer confirmed this.
 |
|
Fig. 63: (a) Electron density profiles of NCAM and PSA-NCAM monolayers bound to lipid monolayers. The peak at ~ 2.5 nm is due to the lipid monolayer. The NCAM box extends 20.6 ± 0.7 nm from the lipid monolayer (blue dash). The PSA-NCAM box extends 29.1 ± 0.5 nm from the membrane in 100 mM NaNO3 (black line), but the box thickness drops to 19.0 ± 0.7 nm in 1 M NaNO3 (red dash). (b) Cartoon (model interpretation) of PSA-NCAM enveloped by swollen PSA in 100 mM salt. (c) Cartoon of PSA-NCAM with collapsed PSA at high salt. |
The thickness of the PSA-NCAM monolayer was substantially greater at 29.1 nm (Figure 63a) [2]. The electron density extends beyond the protein core, and thus shows that the hydrodynamic volume of the polymer chain completely envelops the protein (Figure 63b). This accounts for the dramatic impact of the PSA attachment on both NCAM function and cell adhesion. The extension of the chains normal to the surface also suggests that PSA-NCAM could influence the function of other cell surface adhesion proteins. PSA is charged, so high ionic strength 1M NaCl screens the charges in the polymer backbone, causing the chains to collapse (Figures 63a,c) and the thickness of the protein box to decrease (Figure 63a). The latter finding reveals the structural basis for the salt-dependent increase in adhesion between cells that display PSA-NCAM on their surfaces. PSA collapse reduces the repulsion and the chain extension, enabling NCAM proteins to reform intercellular bonds.
In summary, these reflectivity studies provide unique insights into the structural basis for the modulation of NCAM adhesion by PSA, and the impact of PSA on cell adhesion and associated biological functions.
References
[1] L. Rutishauser, L. Landmesser, Trends Neurosci. 19, 422, (1996).
Principal Publication and Authors
C.P. Johnson (a), G. Fragneto (b), O. Konovalov (c), V. Dubosclard (d), J.-F. Legrand (d) D. Leckband (a,e), Biochemistry, 44, 546 (2005).
(a) Department of Chemistry, University of Illinois (USA)
(b) ILL
(c) ESRF
(d) CEA-CNRS-Universite J. Fourier, Grenoble (France)
(e) Department of Chemical and Biomolecular Engineering, University of Illinois (USA)



