- Home
- Users & Science
- Scientific Documentation
- ESRF Highlights
- ESRF Highlights 2004
- Structural Biology
- The Structure of Bacteriophage Fi12 Packaging ATPase: An RNA Packaging Motor Caught in Action
The Structure of Bacteriophage Fi12 Packaging ATPase: An RNA Packaging Motor Caught in Action
Genome packaging is an essential step in the life cycle of all viruses. Different strategies ranging from co-assembly of the coat protein with the nucleic acid (like in HIV) to active packaging into pre-assembled capsids have been demonstrated. The dsRNA bacteriophage F12 of the Cystoviridae family utilise the latter mechanism. The packaging is performed by a virus motor protein P4 at the expense of ATP hydrolysis. The hexameric packaging motor resides at the five-fold vertices of the procapsid (Figure 76) and serves as the portals for RNA entry. The role of P4 in the packaging of genomic RNA resembles that of the portal complex encountered in dsDNA bacteriophages. While the dsDNA portal complex relies upon a transiently associated terminase complex for ATPase activity, P4 acts as both the ATPase and the portal ring making it one of the simplest viral packaging motors.
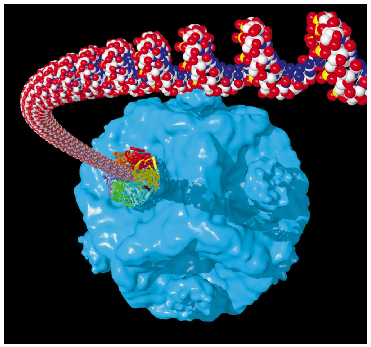 |
Fig. 76: Illustration of the hexameric ATPase P4 in the act of packaging ssRNA into the procapsid of bacteriophage |
In order to better understand how P4 couples ATP hydrolysis to RNA translocation during genome packaging, we have collected data from a selenomethionine-substituted P4 protein crystal in complex with ADP on ESRF beamline BM14 to 2.5 Å resolution and solved the structure by the Multi-wavelength Anomalous Diffraction (MAD) method. The P4 subunit is composed of three domains and belongs to the ![]() /ß -fold family. The central domain of P4 is a Rossmann-type nucleotide binding domain which is structurally similar to that of a large class of hexameric helicases and oligoucleotide translocating enzymes.
/ß -fold family. The central domain of P4 is a Rossmann-type nucleotide binding domain which is structurally similar to that of a large class of hexameric helicases and oligoucleotide translocating enzymes.
To obtain snapshots of the motor protein at different steps on the catalytic pathway we have subsequently generated crystals of ![]() 12 P4 in complex with a panel of different nucleotides (ADP and ATP analogues) and divalent cations. Since the motor is very active, even turning over supposedly non-hydrolysable analogues of ATP at room temperature, some of this work required a careful coordination of low temperature crystallisation and data collection. These structures revealed how ATP, activated by RNA binding, drives RNA translocation through cooperative conformational changes and provided the first insights into the mechanism of active packaging in atomic detail. Furthermore, this provides a model for chemomechanical coupling of this ubiquitous family of proteins of hexameric helicases.
12 P4 in complex with a panel of different nucleotides (ADP and ATP analogues) and divalent cations. Since the motor is very active, even turning over supposedly non-hydrolysable analogues of ATP at room temperature, some of this work required a careful coordination of low temperature crystallisation and data collection. These structures revealed how ATP, activated by RNA binding, drives RNA translocation through cooperative conformational changes and provided the first insights into the mechanism of active packaging in atomic detail. Furthermore, this provides a model for chemomechanical coupling of this ubiquitous family of proteins of hexameric helicases.
Principal Publication and Authors
E.J. Mancini (a), D.E. Kainov (b), J.M. Grimes (a), R. Tuma (b), D.H. Bamford (b) and D.I. Stuart (a), Cell 118, 743-55 (2004).
(a) Division of Structural Biology, The Henry Wellcome Building for Genomic Medicine, Oxford University (UK)
(b) Institute of Biotechnology, University of Helsinki (Finland).



